Figure 1.
Regulatory Element Usage Distinguishes Cell State during Neural Induction
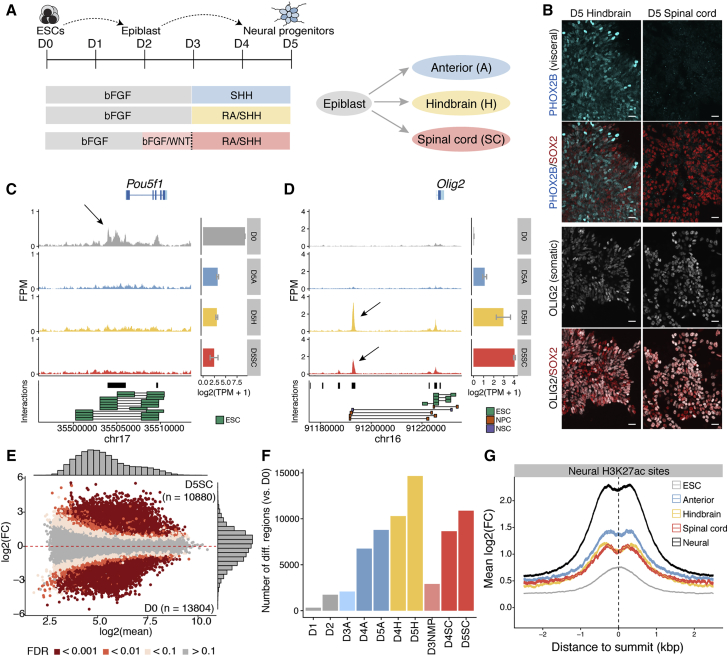
(A) Schematic of mouse ESCs differentiated to NPs with anterior (top), hindbrain (middle), or spinal cord (bottom) identity. Spinal cord progenitors are generated via an NMP state induced by the addition of FGF and WNT signals from day (D) 2 to 3 (light pink shading).
(B) D5 immunofluorescence reveals hindbrain progenitors generate a mixture of PHOX2B expressing visceral and OLIG2 expressing somatic MNs. Spinal cord progenitors lack visceral but generate OLIG2 expressing somatic MN progenitors. Scale bars represent 20 μm.
(C and D) ATAC-seq accessible regions present in ESCs (D0, gray) compared with D5 anterior (D5A; blue), hindbrain (D5H; yellow), and spinal cord (D5SC; red) progenitors and associated gene expression levels determined by mRNA-seq (Gouti et al., 2014; error bars = SEM). cis interactions indicated below each plot represent known genomic interactions from published data (Table S2). ESCs express Oct4 and show accessibility at Pou5f1/Oct4 enhancers (C, arrow). D5H and D5SC NPs have open regions flanking neural expressed Olig2 (D, arrow).
(E) Genome-wide accessibility comparison in D5 spinal cord (D5SC) versus D0 ESCs (false discovery rate [FDR] < 0.01 and |log2(FC)| > 1).
(F) The proportion of differential sites present in each condition compared with ESCs.
(G) Both neural and AP-specific sites, but not ESC sites, are enriched in H3K27ac marks from NPs (Peterson et al., 2012).
bFGF, basic fibroblast growth factor; D, day; ESC, embryonic stem cell; FC, fold change; kbp, kilobase pairs; RA, retinoic acid; SHH, sonic hedgehog; TPM, transcripts per million.