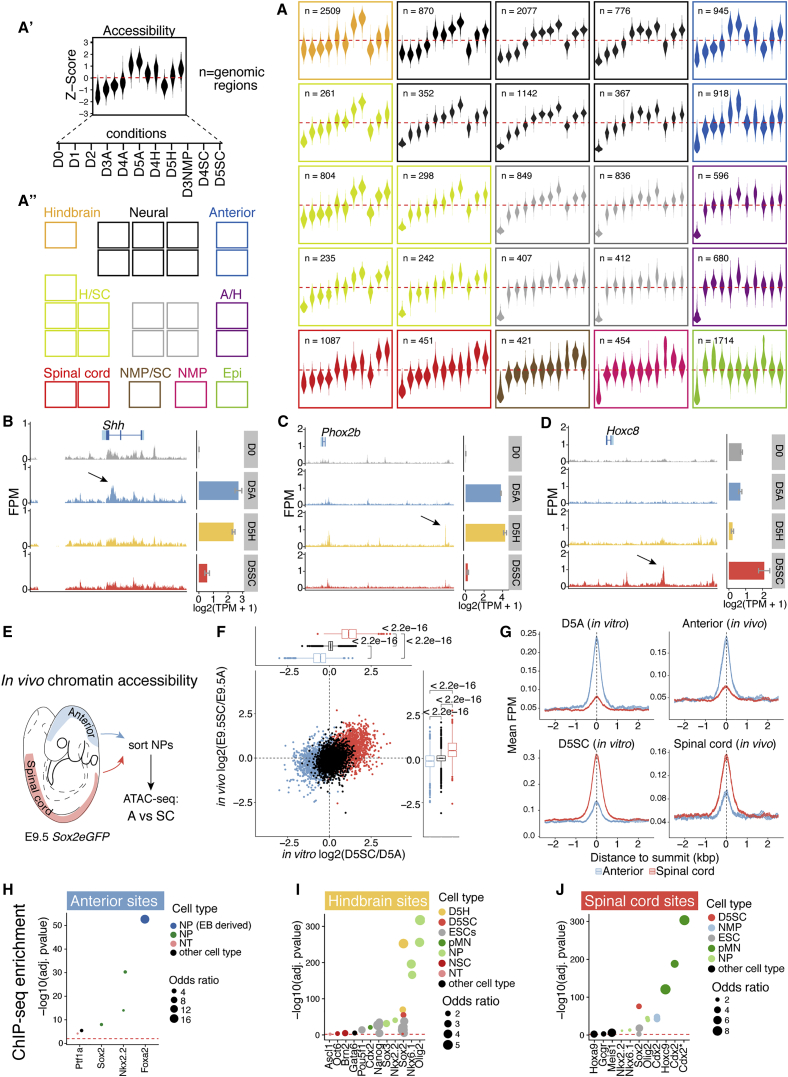
Figure 2.
Differential Enhancer Usage Reveals NP Axial Identity
(A) Self-organizing map (SOM) of regulatory regions showing differential accessibility relative to D0. Each plot represents the chromatin accessibility Z score for each genomic region in the cluster for each condition (see key in A’). Many sites are common (“neural sites”) to all NPs (black cluster; n = 5,584). These differ from epiblast regulatory regions that are accessible at early stages of the differentiation (Epi; green; n = 1,714). Region-specific sites are identified in anterior (blue; n = 1,863), hindbrain (orange; n = 2,509), and spinal cord (red; n = 1,538) progenitors. A distinct set of regulatory regions identifies D3 NMPs (pink; n = 454 regions). A/H represents shared anterior and hindbrain accessible sites (purple; n = 1,276); H/SC, shared hindbrain and spinal cord sites (lime; n = 1,840); and NMP/SC, shared NMP and spinal cord sites (brown; n = 421). Grey are unclassified sites.
(B–D) Examples of ATAC-seq accessible regions that define anterior (B), hindbrain (C), or spinal cord (D) D5 progenitors, identified using the SOM (A). Gene expression from mRNA-seq (error bars = SEM) is shown to the right. Anterior progenitors display region-specific open sites at Shh (B), and hindbrain progenitors demonstrate a Phox2b site (C) and a Hoxc8 site opens in spinal cord (D).
(E–G) In vivo ATAC-seq correlates with in vitro. NPs obtained from brain (E; blue shading) and spinal cord (E; red shading) of E9.5 Sox2eGFP embryos. The fold change in accessibility at anterior (blue; n = 1,863) and spinal cord (red; n = 1,538) sites identified in vitro in spinal cord NPs relative to anterior NPs in vivo correlates with AP identity (F). Common neural sites in vitro (black) are similar in both populations in vivo. Anterior sites identified in vitro show preferential accessibility in vivo in anterior NPs (G), and spinal cord in vitro sites show more accessibility in vivo in spinal cord NPs (p values; Wilcoxon rank-sum test).
(H–J) ChIP-seq enrichment analysis in anterior (H), hindbrain (I), and spinal cord sites (J). SOX2 ChIP-seq in D5 hindbrain (D5H) and spinal cord (D5SC) cells reveals that the binding site preference is condition specific (I and J). CDX2∗ denotes CDX2 ChIP-seq performed in the presence of FGF signaling (Mazzoni et al., 2013).
FPM, fragments per million; neural EB, embryoid bodied-derived NPs; NMP, neuromesodermal progenitors; NP, neural progenitors; NT, neural tube; pMN, motor neuron progenitors. See also Figure S2.