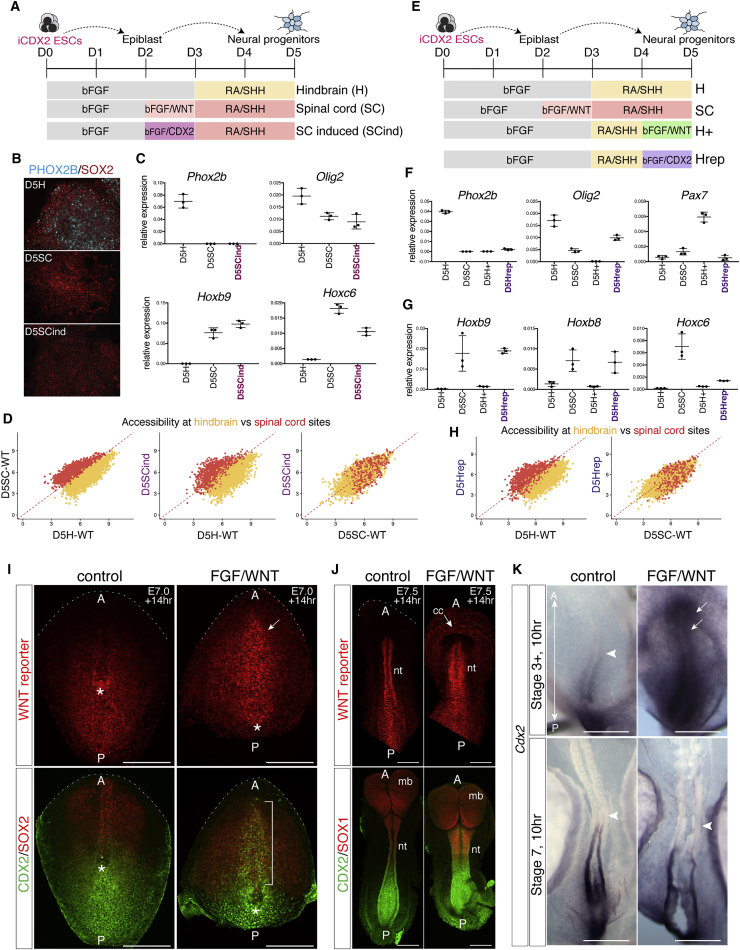
Figure 5.
CDX2 Can Replace WNT and Prolong Spinal Cord Competency
(A) Schematic of the differentiation using iCDX2 ESCs (Niwa et al., 2005) to induce CDX2 between D2 and D3 (SCind).
(B) Immunofluorescence of NPs at D5 PHOX2B (cyan) and SOX2 (red). Cranial MNs in hindbrain, but not spinal cord or SCind.
(C) qRT-PCR analysis at D5 shows that the induction of CDX2 between D2 and D3 maintains Olig2 expression and upregulates Hoxb9 and Hoxc6. Error bars represent the standard deviation.
(D) Chromatin accessibility, measured by ATAC-seq, at hindbrain (yellow) and spinal cord (red) sites at D5. Spinal cord sites are more open in WT spinal cord cells than WT hindbrain cells (left plot). The induction of CDX2 between D2 and D3 (D5SCind) increases accessibility at spinal cord sites versus D5H WT cells (middle plot) and similar levels of accessibility in hindbrain and spinal cord sites when compared to D5SC WT cells (right plot).
(E) Schematic of the differentiation using iCDX2 ESCs to induce CDX2 between D4 and D5 under hindbrain conditions (Hrep) versus hindbrain+ (H+) conditions, in which WNT signaling is activated between D4 and D5.
(F and G) qRT-PCR data indicate that, by D5, the induction of CDX2 is sufficient to repress Phox2b but maintain Olig2 (F), in contrast to H+ cells, which repress Olig2 expression (Figure 3). The induction of CDX2 between D4 and D5 upregulates posterior Hox genes Hoxb9, Hoxb8, and Hoxc6 (G). Error bars represent the standard deviation.
(H) Accessibility at hindbrain (yellow) and spinal cord (red) sites reveals that D5Hrep cells display increased accessibility at spinal cord sites and loss of hindbrain sites compared to D5H cells.
(I and J) WNT reporter embryos cultured for 14 hr in control versus WNT signaling conditions (Table S3) from E7.0 (I) or E7.5 (J). Images show embryos cultured in media containing bFGF (control) versus FGF and CHIR99021 (FGF/WNT) conditions. Embryos are oriented with anterior at the top of the image. Dashed white lines demarcate the anterior limit.
(I) Ventral view of E7.0 cultured embryos. Ectopic induction of WNT activity (n = 10/13) and CDX2 (green, white bracket) is observed in the presence of WNT signaling (n = 10/13), but not control conditions (n = 0/10). SOX2 marks the epiblast (red). Asterisk demarcates the position of the node. Scale bars represent 250 μm.
(J) Ectopic induction of WNT signaling in E7.5 cultured embryos (n = 13/16) versus control conditions (n = 0/6). No CDX2 (green) expansion was detected in the anterior neural plate (n = 0/31) marked by SOX1 (red) versus control conditions (n = 0/30). Top panels in (J) show ventral views; bottom panels in (J) show dorsal views. Scale bars represent 250 μm.
(K) Chick whole-mount in situ hybridization for Cdx2 following 10-hr ex vivo embryo culture from HH stage 3+ (top panels) and stage 7 (bottom panels). The addition of WNT signals promotes ectopic (white arrows) anterior Cdx2 expression at early stages (15/15 in the FGF/WNT versus n = 0/13 control at stage 3+). By contrast, no expansion is observed in response to WNT signaling in stage 7 embryos that already contain a neural plate (n = 0/9 in FGF/WNT and n = 0/12 in control). White arrowheads mark the anterior limit of Cdx2 expression. Scale bars represent 500 μm.
cc, cardiac crescent; mb, midbrain; nt, neural tube; p, posterior.