Figure S1.
Quality Control for All ATAC-Seq Samples Generated in This Study, Related to STAR Methods and Figure 1
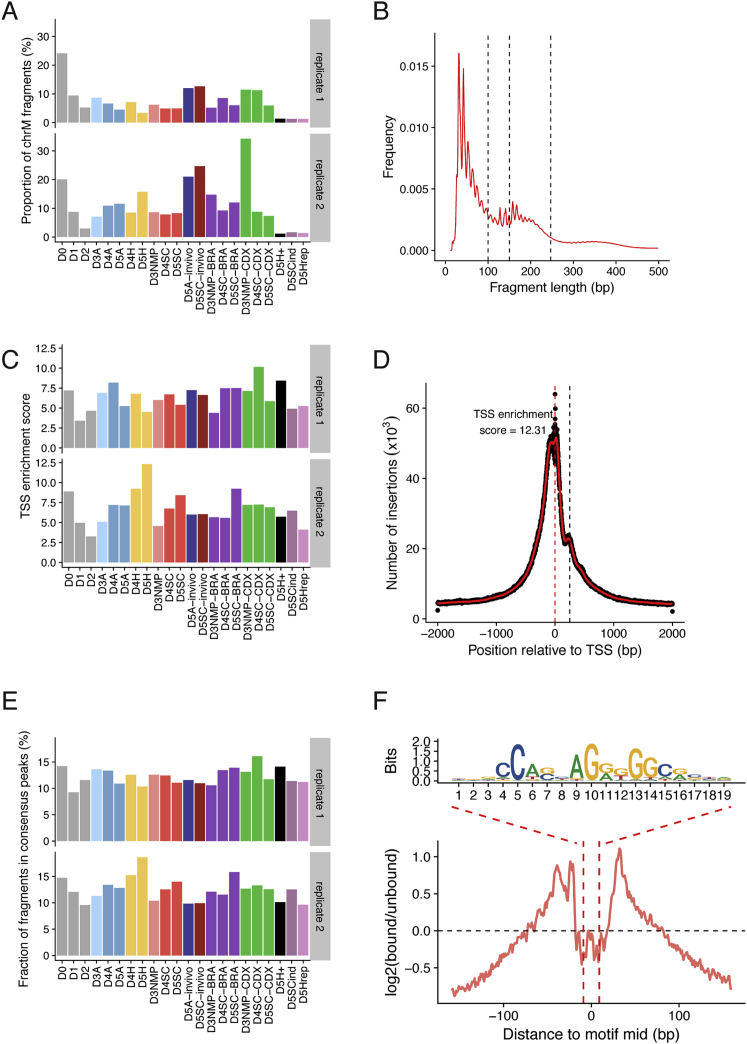
(A) The proportion of mitochondrial fragments recovered across each sample.
(B) Representative example showing the distribution of fragment lengths recovered from ATAC-seq, using paired-end sequencing.
(C) Average level of Tn5 enrichment (score = maximum(number of insertions)/minimum(number of insertions)) observed across transcription start sites (TSS) for each sample.
(D) Summarized Tn5 insertion profile covering ± 2kb around annotated TSS for sample D5H (replicate 2). Red line corresponds to a 50bp running average.
(E) The fractions of fragments that map to in vitro consensus peak regions.
(F) CTCF footprint present in ESC accessible regions as determined by ATAC-seq.