Figure S4.
Expression Dynamics of Cdx TFs during the Spinal Cord Differentiation, Related to Figure 4
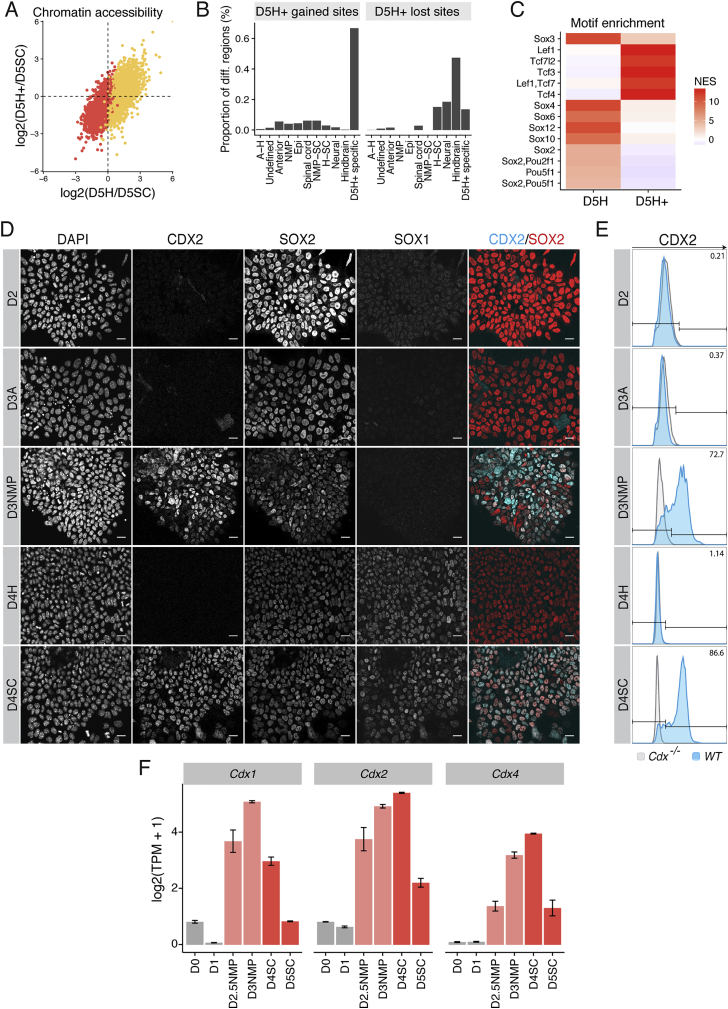
(A) Scatterplot showing the fold change of D5H+ or D5H cells relative to the D5SC condition. Note that the majority of hindbrain sites (yellow) remain accessible in D5H+ treated cells, in contrast to spinal cord (red) sites.
(B) The distribution of genomic regions, as defined in the self-organizing map (SOM; see Figure 2A), in D5H+ cells that show changes in accessibility when compared to D5H cells (FDR < 0.01 & |log2(FC)| > 1). Note that D5H+ cells do not gain sites associated with D3NMP, which are treated with WNT between D2-3 (see columns labeled “NMP”), but gain additional sites not classified in the SOM (see columns labeled “D5H+ specific).
(C) Motif enrichment using iCis Target (Imrichová et al., 2015) on genomic regions that are differentially accessible between D5H and D5H+ reveals an enrichment of TCF/Lef factors following WNT treatment in D5H+ cells, in contrast to the D5H condition which harbours sites enriched in SOX factors.
(D) Confocal microscopy of hindbrain and spinal cord cells from D2-4 shows the induction of CDX2 at D3 in FGF/WNT in the spinal cord condition (D3NMP). SOX1 expression, marking neural progenitors, is not detected until D4 in both conditions, following RA and SHH treatment. Right column shows CDX2/SOX2 composites.
(E) Flow cytometry in WT (blue) versus Cdx triple mutant ESCs (grey) at the indicated time points and conditions indicates the percentage of SOX2 positive cells that express CDX2.
(F) Expression profile determined by mRNA-seq (Gouti et al., 2014) for Cdx1, Cdx2 and Cdx4 from D0 to D5 of the spinal cord differentiation. Error bars = SEM. NMP=neuromesodermal progenitor; SC=spinal cord; TPM=transcripts per million.