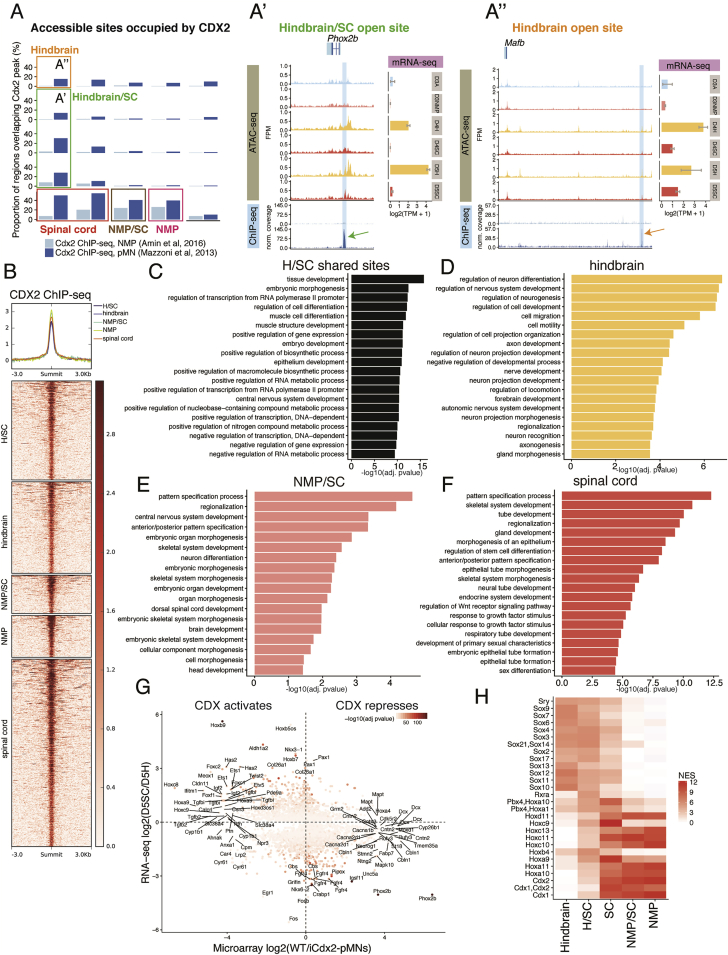
Figure S5.
CDX2 Occupancy in Open Chromatin Sites and Associated Gene Ontology Enrichment, Related to Figure 4
(A) The proportion of accessible regions bound by CDX2, as indicated by CDX2 ChIP-seq analysis from neuromesodermal progenitors (NMP, light blue bars, Amin et al., 2016) and motor neuron progenitors (pMNs, dark blue bars, Mazzoni et al., 2013) derived in vitro, compared with the accessible regions recovered from the self-organizing map (SOM) in this study (refer to Figure 2A). The overlap demonstrates that CDX2 binds to NMP, NMP/SC (NMP and spinal cord shared) and spinal cord (SC) sites identified by ATAC-seq. Furthermore, in pMN conditions, CDX2 binds accessible regions that are shared between the hindbrain and spinal cord (A’, boxed region outlined in green). CDX2 also targets hindbrain accessible sites (A’’). (A’) The Phox2b genomic region represents a shared hindbrain/spinal cord accessible site that is bound by CDX2 in pMN conditions. (A’’) A hindbrain-accessible site is bound by CDX2 at the Mafb locus in pMN conditions.
(B) Region heatmap showing CDX2 binding at open chromatin sites recovered from the SOM (pMN; Mazzoni et al., 2013).
(C–F) Gene ontology enrichment analysis for CDX2-bound regions shown in (B). In hindbrain accessible regions (D), CDX2 binding is associated with neural genes in contrast to either the NMP and spinal cord (NMP/SC) shared (E) or SC-specific sites (F), which target genes involved in anterior-posterior patterning.
(G) Comparison of log2 fold gene expression changes in D5 spinal cord (D5SC) versus D5 hindbrain (D5H), determined by mRNA-seq (Gouti et al., 2014), and wild-type (WT) versus Cdx2-induced motor neuron progenitors (iCdx2-pMNs) determined by microarray (Mazzoni et al., 2013). CDX2 induction positively correlates with Hoxb9 and other 5′ Hox genes while it negatively correlates with Aldh1a2 in the spinal cord, in agreement with previous studies (Gouti et al., 2017). CDX negatively correlates with hindbrain genes including Phox2b. Color filling indicates –log10(adj. pvalue) from the D5SC versus D5H comparison using DESeq2 (Love et al., 2014).
(H) Motif enrichment analysis of CDX2-bound regions shown in (B). Heatmap colors represent the normalized enrichment score computed using iCis Target (Imrichová et al., 2015). CDX2 binds to hindbrain accessible regions that are enriched with SOX factor motifs, in contrast to HOX motifs found in spinal cord sites.