Figure 3.
Localization of the PCGF2 Mutations
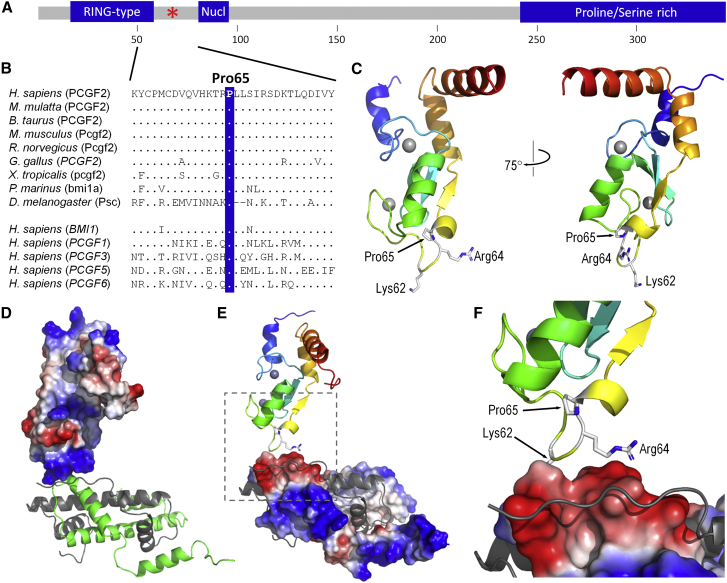
(A) Schematic domains of PCGF2. The domains and motifs of PCGF2 (UniprotKB: P35227) are illustrated. These include a RING-type Zinc finger (residues 18–57), nuclear localization signal (81–95) and Proline/Serine-rich domain (242–344). The location of the Pro65 residue is marked by the red asterisk. Residue number is indicated in the scale below the illustration.
(B) The PCGF2 mutations are located at the highly conserved Pro65 residue. ClustalW homology alignments for Human PCGF2 (residues 51-80) and a range of orthologs and paralogs. Orthologs include Human (NP_009075.1), Rhesus monkey (XP_001083817.1), Cow (NP_001137578.1), Mouse (NP_001156779.1), Rat (NP_001099306.1), Chicken (XP_003642857.1), Frog (NP_001025573.1), Lamprey (ENSPMAP00000007297.1), and Fruit fly (NP_523725.2). Paralogs include BMI1/PCGF4 (NP_005171.4), PCGF1 (NP_116062.2), PCGF3 (NP_006306.2), PCGF5 (NP_001243478.1), and PCGF6 (NP_001011663.1). Identical residues are indicated by dots. The blue bar highlights the position of the Pro65 residue.
(C) Structural model of human PCGF2 (residues 5-101, modeled on template 2h0d). Two views of the model are shown in ribbon format, colored from blue, N-terminal, to red, C-terminal, with side-chains shown for Pro65, Arg64, and Lys62. Grey spheres represent bound zinc ions; interaction with RING1B is primarily mediated via residues in helices 1 and 3 (blue and orange, respectively).
(D) The interaction between PCGF2 and histone H3/4 (modeled on template 4r8p). The predicted molecular surface of PCGF2 (left) is colored by electrostatic charge (blue, basic; red, acidic); histone chains from 4r8p are shown as green (H3.2) and gray (H4) ribbons, respectively; other chains of the complex have been omitted for clarity; note the basic patch of PCGF2 in contact with H3.2.
(E) As (D), but showing surface charge for H3.2, with PCGF2 shown as a ribbon colored from N-terminal, blue to C-terminal, red; note the acidic patch of H3.2 lying opposite the basic patch of PCGF2.
(F) As (E), but showing detail around the PCGF2/H3.2 interface; the regions shown are outlined by gray broken lines in part E; sidechains of PCGF2 Pro65, Arg64, and Lys62 (partially obscured) are shown in stick format.