Figure 1.
Family Pedigrees, Electropherograms of Identified ATP1A1 Mutations, and Localization of Mutations Within the ATP1A1 Protein (α1 Subunit of Na+, K+-ATPase) with Multiple Sequence Alignment
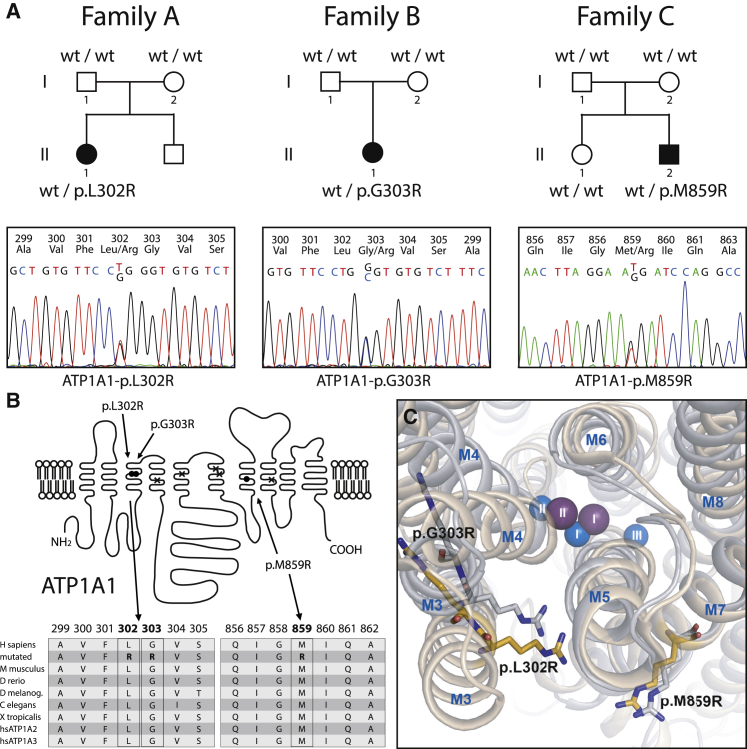
(A) Heterozygous ATP1A1 mutations p.Leu302Arg (p.L302R), p.Gly303Arg (p.G303R), and p.Met859Arg (p.M859R) identified in the three individuals (A-II-1, B-II-1, and C-II-2) were not present in unaffected parents but occurred de novo.
(B) Whereas adjacent amino acid residues Leu302 and Gly303 are located within the third transmembrane domain, M859 lies within the seventh transmembrane helix of the encoded α1 subunit of Na+, K+-ATPase (filled circles). Crosses indicate ion-binding carboxylate residues (Glu334 in M4; Glu786 in M5; Asp811 and Asp815 in M6; and Asp933 in M8). A multiple-sequence alignment (RefSeq NP_000692, UniProt P05023) of ATP1A1 amino acid residues surrounding mutated positions p.302, p.303, and p.859, respectively (bold), is shown. All three positions are highly conserved between species and between α subunit homologs ATP1A1, ATP1A2, and ATP1A3.
(C) Structural location of the arginine residues in mutants p.Leu302Arg, p.Gly303Arg, and p.Met859Arg. The arginines were inserted into the atomic models derived from crystal structures with Na+ or K+ bound. The central transmembrane domain of the α1 subunit, consisting of helices M3 to M8 with bound ions (Na+ blue, K+ purple, numbering according to conventional nomenclature) is shown as seen from the extracellular surface. The bulky arginines seem to be able to disturb the ion-binding sites I and II. The p.Gly303Arg arginine is predicted to collide with M4, whereas the p.Leu302Arg arginine most likely collides with M5, particularly in the K+-bound form. Finally, the p.Met859Arg arginine might interact with an M7 glycine essential to the M7 kink that makes room for the binding of the K+ ions.