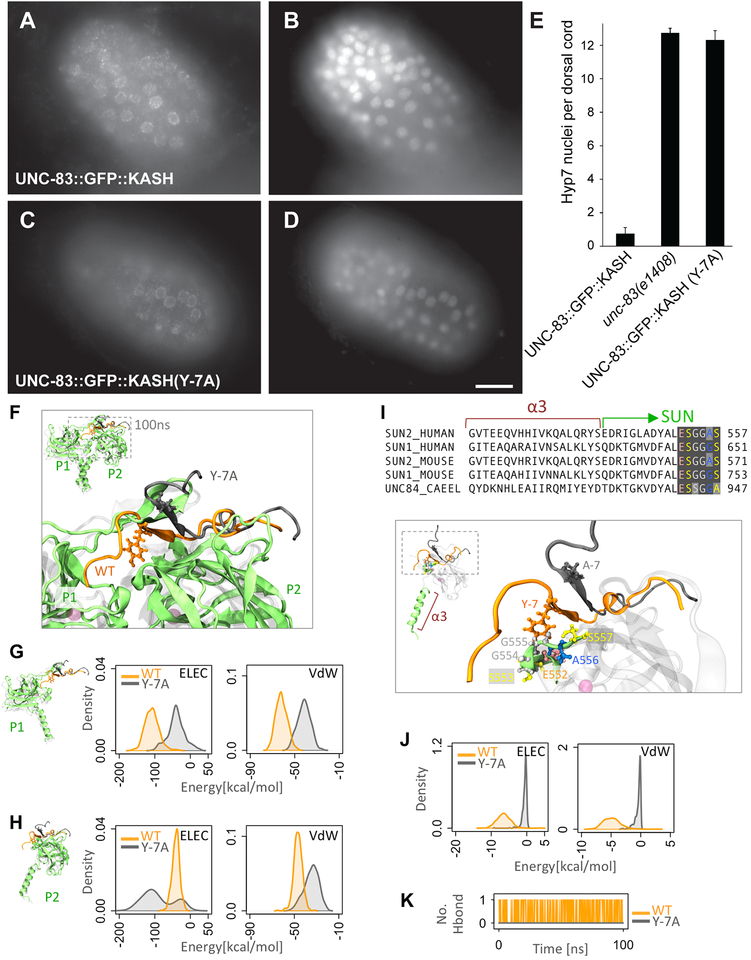
Figure 3. Mutation of tyrosine −7 of KASH disrupts SUN-KASH interactions.
(A,C) Anti-GFP immunolocalization and (B,D) DAPI staining in pre-bean embryos around the time of hyp7 nuclear migration. (E) Quantification of nuclear migration defects as in Fig 2. Error bars are 95% CI; scale bar is 10 μm. (F-K) Modeling the effect of Y-7A on SUNKASH interactions. (F) Superimposed structures of wild type (orange) and Y-7A (gray) after 100ns of molecular dynamics simulation time. Only two of three SUN2 protomers are shown (P1 and P2). (G-H) Electrostatic (ELEC) and Van der Waals (VdW) energies between KASH (residues 0 to −17) and (G) the P1 or (H) P2 SUN2 protomer for WT (orange) and Y-7A (gray) (shown as densities over 100ns simulation time). (I) Sequence and structure of α3 and first few residues of the SUN domain showing that the major interacting partners of Y-7 on SUN P2 (E552-S557) are highly conserved. (J) ELEC and VdW energies between residue −7 and residues E552-S557 on SUN2 P2 for WT (orange) and Y-7A (gray) (shown as densities over 100ns simulation time). (K) Number of H-bonds between residue −7 and residues E552-S557 on SUN2 P2 for WT (orange) and Y-7A (gray).