Abstract
Preterm birth, or the delivery of an infant prior to 37 weeks of gestation, is a significant cause of infant morbidity and mortality. In the last decade, the advent and continued development of molecular profiling technologies has enabled researchers to generate vast amount of ‘omics’ data, which together with integrative computational approaches, can help refine the current knowledge about disease mechanisms, diagnostics, and therapeutics. Here we describe the March of Dimes’ Database for Preterm Birth Research (http://www.immport.org/resources/mod), a unique resource that contains a variety of ‘omics’ datasets related to preterm birth. The database is open publicly, and as of January 2018, links 13 molecular studies with data across tens of thousands of patients from 6 measurement modalities. The data in the repository are highly diverse and include genomic, transcriptomic, immunological, and microbiome data. Relevant datasets are augmented with additional molecular characterizations of almost 25,000 biological samples from public databases. We believe our data-sharing efforts will lead to enhanced research collaborations and coordination accelerating the overall pace of discovery in preterm birth research.
Subject terms: Reproductive disorders, Data integration, Data publication and archiving, Neonatology
Background & Summary
In the last decade, the advent and continued development of genotyping and next-generation sequencing technologies has enabled researchers to generate a vast amount of molecular data. The current and ever-growing availability of public ‘omics’ databases, including Gene Expression Omnibus1,2, Array Express3, dbGAP4,5, and numerous other repositories, along with computational tools to reveal molecular drivers of disease at a network level, present a unique and new opportunity to refine current knowledge about disease mechanisms, diagnostics, and therapeutics. In addition, novel technologies are allowing high-throughput measurements of the human genome, epigenome, transcriptome, and immunome at the population level6. There have been several disease- and phenotype-specific efforts to aggregate datasets and to make them available in the public domain, with exemplary strides by the cancer community including The Cancer Genome Atlas (TCGA)7, which captures ‘omics’ profiling of tens of thousands of cancer samples; Cancer Cell Line Encyclopedia (CCLE), which focuses on molecular profiling of cancer cell lines; the Library of Integrated Network-Based Cellular Signatures (LINCS)8, which focuses on transcriptomic profiling of small molecules and other perturbations in cancer lines; and many others. However, other fields are lagging behind in the data-sharing realm. To this end, we aim to enable open data-sharing efforts in the fields of obstetrics and gynecology. Here, we present our March of Dimes (MOD) Database for Preterm Birth Research (http://www.immport.org/resources/mod), a unique resource that captures a variety of ‘omics’ datasets related to preterm birth (PTB).
PTB, or the delivery of an infant prior to 37 weeks of gestation, is a significant cause of infant morbidity and mortality. Globally, approximately 11% of infants are born prematurely every year, totaling nearly 15 million births. Infants born preterm are at risk for a variety of adverse outcomes, such as respiratory illness, blindness, and cerebral palsy, with associated complications resulting in nearly one million deaths each year9–11. Despite many attempts for preterm birth prevention, there is still an acute problem with prevalence rising according to the World Health Organization. Spontaneous preterm birth, accounting for two thirds of all preterm births, is considered a complex phenotype with no single known cause12 or biological basis13,14. One mechanism that has been associated with spontaneous preterm birth is chorioamnionitis, a condition associated with microbial infection of the amniotic fluids. Other suggested causes of PTB are progesterone deficiency, cervical insufficiency, disruption of the immune tolerance of the mother towards the fetus and disruption of the vaginal microbial balance, causing an inflammatory process15–19. Because of the complexity of the phenotype, a comprehensive integrative approach is needed to better understand the etiology of preterm birth and inform new diagnostic and therapeutic strategies.
The March of Dimes is dedicated to not only helping affected infants and families; but also preventing PTB. Only modest progress has been made in identifying the underlying causes of PTB, so the MOD has made this a top research priority. To foster a new model of collaboration with the hope of leading to transformative discoveries, six MOD Prematurity Research Centers have been launched. The goals are to integrate scientists from individual disciplines and to form innovative collaborations that can accelerate research discoveries20. The first Center was launched in 2011 at Stanford University School of Medicine to study infection and inflammation, transcriptomics and lead bioinformatics effort in PTB research. The second center, the MOD Prematurity Research Center - Ohio Collaborative, was opened in 2013 with a focus on evolutionary biology and genetics in PTB as well as molecular development of pregnancy, progesterone signaling and racial disparities in PTB. In 2014, two more centers were launched. On November 10, 2014 the MOD Prematurity Research Center at Washington University was established in St. Louis to apply bioengineering to study cervical remodeling and uterine contractility, as well as determining if chronodisruption is a risk factor for preterm birth. On November 17, 2014 the MOD Prematurity Research Center at the University of Pennsylvania was opened to address questions of cervical remodeling, placental development and bioenergetics in the context of PTB. The fifth center, MOD Prematurity Research Center University of Chicago-Northwestern-Duke was launched in 2015 with a focus on what causes premature birth, and specifically address six interrelated transdisciplinary research themes around gene regulation. The most recent center was launched in February 2018 at the Imperial College London with a focus on how the body recognizes and interacts with bacteria and other microbes in the birth canal that may increase the risk of premature birth.
There have been several previous efforts to aggregate and integrate genomics data for PTB research21. dbPTB22 is a resource that collects genomic data across a large number of studies focusing on linking information from published literature with data from expression databases, linkage studies, and pathway analyses to identify biologically relevant genes for testing in an association study of genetic variants and PTB. However, the database is limited to disease-gene and pathway associations. GEneSTATION23 is a comprehensive database that integrates cross-species genomic, transcriptomic, and evolutionary data to advance the understanding of the genetic basis of gestation - and pregnancy-associated phenotypes and to accelerate the translation of discoveries from model organisms to humans. This resource specifically focuses on evolutionary and genomic data and is not designed for capturing other ‘omics’ technologies that cannot be directly mapped to human genes or genetic elements (e.g., microbiome data). In addition, dbPTB does not allow for bioinformatics re-analyses of the processed datasets they aggregate and only summarizes the data at the results level; whereas, GEneSTATION is designed for exploratory data analyses and not as a tool for large-scale re-analyses or meta-analyses of pregnancy ‘omics’ data in general. A comprehensive resource that can capture more diverse types of data and enable data re-use and re-analyses is needed.
The MOD Database for Preterm Birth Research aims to organize scientific and clinical research data across the six MOD-Funded Prematurity Research Centers with the goal of enhancing research collaborations and accelerating the overall pace of discoveries in this field. Data from the Centers includes a diverse set of processed ‘omics’ data and results files as well as data generation protocols to support re-analyses and meta-analyses of the datasets. As of January 2018, the database references 9 studies across over 350 patients and with individual level molecular data on nearly 8,000 samples from 6 measurement modalities. Four additional large-scale GWAS studies across tens of thousands of patients are also included, for which only summary-level data are available. The repository includes genomic, transcriptomic, immunonological, and microbiome data that are available freely to the scientific community. Having all the data aggregated as part of the same resource, substantially extends the value the data allowing researchers to integrate the information and ask novel research questions.
Methods
The database development was undertaken in collaboration with Northrop Grumman (NG) – Health Solutions, partner of the National Institute of Allergy and Infectious Diseases (NIAID) Division of Allergy, Immunology, and Transplantation (DAIT). Since 2004, NG has been the prime contractor for the ImmPort24 database and data-sharing portal, working with researchers at UCSF and Stanford to ensure that NIAID-funded discoveries serve as the foundation of future research. In order to host the MOD Database for Preterm Birth Research, we used the existing infrastructure, data model, and repository schema, known as ImmPort (www.immport.org)24,25. ImmPort encompasses immunological data across a wide range of diseases and conditions including several pregnancy-related studies. Study data includes over 50 diverse types of data including arrays, mass cytometry (CyTOF), enzyme-linked immunosorbent assays (ELISA), flow cytometry, and gene expression as well as others.
Datasets archived in ImmPort cover a broad spectrum of study-specific data including protocol designs, assay protocols, treatment and sampling time points, and subject demographics. Datasets are curated and organized by the ImmPort team prior to sharing with the scientific public. Curation efforts include adoption of community standards and controlled vocabularies across a broad spectrum of variables to improve data consistency within and across studies facilitating data re-use. Data upload templates instantiate standards and ontologies and are the result of ongoing outreach with domain experts, standards working groups, and research consortia. Data are de-identified. ImmPort has adopted best-practices in human study participant de-identification such that Health Insurance Portability and Accountability Act of 1996 (HIPAA)-restricted data are not captured in ImmPort. In addition, ImmPort references genetics data stored within the National Center for Biotechnology Information (NCBI) Database of Genotypes and Phenotypes (dbGaP), ensuring ImmPort data safeguards potentially identifying information.
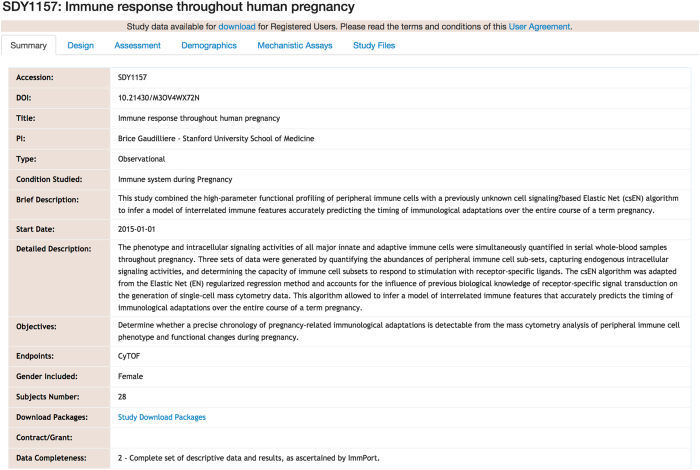
For each study (Fig. 1), a summary page is created, which contains the description, principal investigators (PIs), links to the publication, and external repositories with raw data. Only processed ‘omics’ data are stored in the repository to avoid duplication of effort. The information is drawn from the publication when available and confirmed by the PIs. Study design, types of assessments, and mechanistic assays with links to the protocols as well as the processed files are listed as separate tabs for each study. Finally, demographics are aggregated across all the samples for each study cohort where individual level data are available. Each subject is given a unique ID and when data from multiple assays is available for the same subject, the data can be accessed by the subject ID. All the human subjects work has been approved by the relevant IRB boards at each institution.
Figure 1. Screenshot of an example study indexed in the resource (SDY1157).
Data Records
As of January 2018, the resource indexed 13 studies (Data Citations 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13), with individual level molecular measurements on nearly 8,000 samples from more than 350 patients, capturing data from 6 measurement modalities (Fig. 2). The resource is further divided into three sections: 1) “curated datasets”; 2) “research highlights”; and 3) “other relevant publicly-available resources relevant to PTB research”. The resource is updated at least on a quarterly basis as new studies get added.
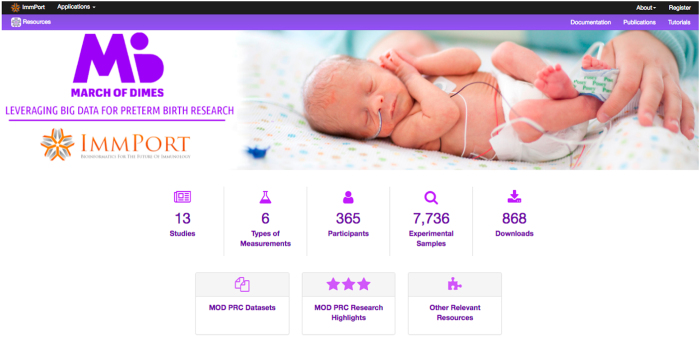
Figure 2. The MOD Database for Preterm Birth Research.
A screenshot of the data repository resource as of January 2018. The landing page contains repository statistics, including the number of studies, participants, experimental samples, and study downloads. There are three sections including: 1) a table of the studies with corresponding links; 2) research highlights listing a selection of recent publications with links to the studies; and 3) a compilation of other relevant publicly-available datasets.
The studies referenced by the database (Table 1) cover a diverse set of modalities including microbiome, CyTOF, RNA-Seq, cell-free DNA and RNA sequencing, and genotyping. The protocols for data generation are included in each study (Fig. 1), as are subject demographics and relevant clinical variables as well as processed ‘omics’ datasets. When possible, raw data stored in other public repositories such as SRA, dbGaP and GEO are linked. Publications arising from the studies are referenced on individual study pages, and the most recent papers are also listed on the “research highlights” section of the resource.
Table 1. MOD-funded studies in the MOD database for preterm birth research as of January 2018.
| ID | Type of Data | Patients | Samples | Principal Investigator | Center | Unprocessed Data | Processed Data | PubMed ID |
|---|---|---|---|---|---|---|---|---|
| *No individual level genomic data is available, only summary statistics and results. | ||||||||
| SDY465 (Data Citation 1) | Microbiome 16S | 47 | 4,122 | David Relman | Stanford University | Raw reads Link to SRA | .BIOM format | 2628335726 |
| SDY475 (Data Citation 2 | CYTOF | 23 | 95 | Martin Angst | Stanford University | 3,000,000 cells per sample | Cell Clusters | 2619006327 |
| SDY775 (Data Citation 3) | Microbiome 16S | 7 | 69 | Samuel Parry and Frederic Bushman | University of Pennsylvania | Raw reads | .BIOM format | 2733872828 |
| SDY1155 (Data Citation 4) | RNA-Seq | 15 | 75 | Catalin Buhimschi | Ohio Collaborative | Sequencing | Counts matrix | 2745243529 |
| SDY1164 (Data Citation 6) | Microbiome 16s | 136 | 2,177 | David Relman | Stanford University | Raw reads | .BIOM | 2884794130 |
| SDY1157 (Data Citation 7) | CYTOF | 28 | 112 | Nima Aghaeepour and Brice Gaudilliere | Stanford University | 3,000,000 cells per sample | Cell clusters | 2886449431 |
| SDY673 (Data Citation d8) | Cell Free DNA and RNA Seq | 50 | 188 | Stephen Quake | Stanford University | ~20,000,000 reads per sample | Counts matrix | 2890405632 |
| SDY1175 (Data Citation 8) | Cell Free DNA Seq | 16 | 58 | Stephen Quake | Stanford University | ~20,000,000 reads per sample | Abundance matrix | 2883099933 |
| SDY776* (Data Citation 9) | Genotyping | 1,705 | 1,705 | Louis Muglia | Ohio Collaborative | Genotypes Link to dbGaP | Association test results | 2628479034,35 |
| SDY1173* (Data Citation 10) | Genotyping | ~40k | ~40k | Louis Muglia | Ohio Collaborative | Genotypes Link to dbGaP | Association test results | 2887703135 |
| SDY1206* (Data Citation 11) | Microbiome | 77 | 149 | Molly Stout | Washington University in St. Luis | Raw reads link to Short Read Archive (SRA) | Abundance matrix | 2854998136 |
| SDY1215* (Data Citation 12) | Genotyping (Mitochondria) | ~15k | ~15k | Neal Sondheimer | University of Pennsylvanya in collab with Stanford | Genotypes Link to dbGaP | Association test results | 2924952337 |
| SDY1205* (Data Citation 13) | Genotyping | ~15k | ~15k | Marina Sirota and Atul Butte | Stanford in collab with Ohio Collaborative | Genotypes Link to dbGaP | Association test results | 2931770138 |
Publicly-available data are displayed in a table format, which currently lists 15 additional studies from GEO, dbGaP, SRA, and ImmPort (Table 2). Various types of transcriptomic, microbiome, and genomic data across a variety of relevant reproductive tissues are included in the database with molecular profiles for more than 25,000 samples. The studies are linked from the resource page to their respective study detail pages in their respective repositories.
Table 2. Other publicly available studies relevant to preterm birth as of January 2018.
| Study ID | Database | Type | Number of Samples | Tissue |
|---|---|---|---|---|
| *No individual level genomic data is available, only linking out to the relevant resources. | ||||
| GSE46510 | GEO | Transcriptomics | 154 samples | Maternal Blood |
| GSE59491 | GEO | Transcriptomics | 326 samples | Maternal Blood |
| GSE73685 | GEO | Transcriptomics | 183 samples | Amnion, Cord Blood, Decidua, Maternal Blood |
| phs001320.v1.p1 | dbGaP | Transcriptomics | 58 samples | Chorionic villus sampling |
| PRJNA242473 | SRA | Microbiome | 349 samples | Vaginal Microbiome |
| phs000735.v1.p1 | dbGaP | Microbiome | 48 samples | Placenta |
| phs000256.v3.p2 | dbGaP | Microbiome | 3,474 samples | Vaginal Microbiome |
| phs000332.v2.p2 | dbGaP | Genetics | 1,779 samples | Blood |
| phs000714.v1.p1 | dbGaP | Genetics | 2,928 samples | Blood |
| phs000353.v1.p1 | dbGaP | Genetics | 3,478 samples | Blood |
| phs000103.v1.p1 | dbGaP | Genetics | 2,000 mother-child pairs | Blood |
| phs001055.v1.p1 | dbGaP | Genetics | 2,397 samples | Blood |
| phs000276.v2.p1 | dbGaP | Genetics | 5,415 samples | Blood |
| SDY37 | ImmPort | ELISA, Cytokines | 55 samples | Maternal Blood |
| SDY36 | ImmPort | Vaccine Response | 300 samples | Maternal Blood |
Technical Validation
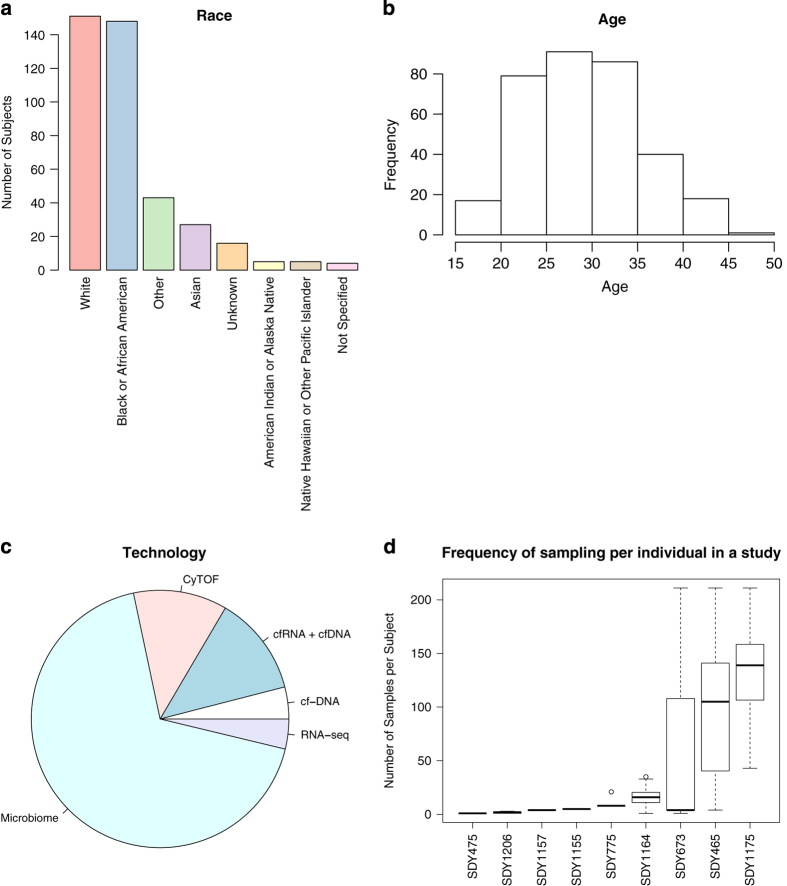
In order to technically validate the resource, we computed some overall statistics on the repository (Fig. 3) focusing on the studies where we had individual level data (as opposed to aggregated summary statistics). As of January 2018, the database cohort consisted primarily of samples from White and African-American individuals with smaller proportions of samples from Asian and other racial and ethnic groups (Fig. 3a). The age of participants that we have individual level data on ranged from 16 to 46 years of age with a mean of 29.7±6.1 years (Fig. 3b). The majority of individuals (67.9%) in the resource have been profiled with microbiome 16s technology (Fig. 3c). There is a significant proportion of cell-free DNA and cell-free RNA measurements, as well as CyTOF (12.5 and 11.8% respectively). While the majority of the studies are cross-sectional and have very few samples per individual, there are several longitudinal studies (SDY465, SDY763 and SDY1175) with extensive sampling frequency (Fig. 3d).
Figure 3. Overall Database Statistics on Individual Measurements (9/13 studies).
(a) Race distribution across the cohort. (b) Age distribution across the cohort. (c) Percentage of samples profiled by each technology from the individual measurements. (d) Frequency of sampling per individual in each study.
We have also been tracking downloads through ImmPort for individual studies as well as bulk downloads as part of the ALLSTUDIES package, which contains the entirety of publicly-available ImmPort studies (Table 3). Individual study downloads counts range from 0 to 61 downloads with an average of 20 downloads per study in addition to a total of 666 downloads of the whole repository as of January 2018.
Table 3. Download statistics for each study as of January 2018.
| Study | First Data Release | Date of First Data Release | Number of downloads as of 01/22/2018 |
|---|---|---|---|
| SDY465 | DR18 | March 18, 2016 | 41 |
| SDY475 | DR16 | December 14, 2015 | 54 |
| SDY673 | DR24 | November 7, 2017 | 5 |
| SDY775 | DR20 | January 31, 2017 | 27 |
| SDY776 | DR24 | November 7, 2017 | 1 |
| SDY1155 | DR23 | September 29, 2017 | 8 |
| SDY1157 | DR23 | September 29, 2017 | 13 |
| SDY1164 | DR23 | September 29, 2017 | 40 |
| SDY1173 | DR24 | November 7, 2017 | 5 |
| SDY1175 | DR24 | November 7, 2017 | 8 |
| SDY1205 | DR25 | January 4, 2018 | 61 |
| SDY1206 | DR25 | January 4, 2018 | 0 |
| SDY1215 | DR25 | January 4, 2018 | 0 |
| Total | 263 | ||
| Bulk Download | 666 |
Usage Notes
The overarching goal of our data-sharing effort is to enable new scientific discoveries from the rich molecular resources that have been funded by the MOD to advance the research in PTB. By making all the data publicly available, we aim to engage the larger research community in tackling this important problem. We are also working on incorporating other types of data into this repository including activity monitoring, sociobehavioral data, imaging and others. In addition to the traditional ‘omics’, investigators are studying non-coding regulatory regions, role of mitochondria, patient-specific glycans and sleep-wake cycles in the context of birth timing. We hope our efforts will entice industry leaders in the areas of machine learning and artificial intelligence to develop and apply computational methodologies to PTB research by leveraging the data that we aggregate. Our group has had tremendous success in mining publicly-available datasets to enable computational drug discoveries in the areas of autoimmunity and cancer39–45. More recently, we have carried out an integrative ancestry-specific GWAS analysis in PTB by integrating internal- and publicly-available data and identified several variants associated with early PTB38 as well as a transcriptomics meta-analysis of maternal and fetal signals elucidating the role of the immune system in parturition timing46. We will continue similar efforts in the area of PTB, and we hope to inspire others to follow suit. We hope this work enabling intersection of data layers related in the context of preterm birth will lead to discovery of novel diagnostic biomarkers and ultimately aid in formulating more effective interventional strategies for the management and prevention of PTB.
Additional information
How to cite this article: Sirota, M. et al. Enabling precision medicine in neonatology, an integrated repository for preterm birth research. Sci. Data. 5:180219 doi: 10.1038/sdata.2018.219 (2018).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases (Bioinformatics Support Contract HHSN272201200028C) and the March of Dimes. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the March of Dimes.
Footnotes
R.L. and J.L.S. are employees of the March of Dimes, which is the funding organization of this project. Since they have contributed to the resource design and inception, they are co-authors on this work.
Data Citations
- DiGiulio D. B., et al. . 2016. ImmPort. https://doi.org/10.21430/M3D491LGDT
- Gaudilliere B., et al. . 2015. ImmPort. https://doi.org/10.21430/M3D8CS7ILY
- Lauder A. P., et al. . 2017. ImmPort. https://doi.org/10.21430/M3PZM1ERD2
- Ackerman W. E., et al. . 2017. ImmPort. https://doi.org/10.21430/M34I5YT3K9
- Callahan B. J., et al. . 2017. ImmPort. https://doi.org/10.21430/M37W3869AH
- Aghaeepour N., et al. . 2017. ImmPort. https://doi.org/10.21430/M3OV4WX72N
- Pan W., et al. . 2017. ImmPort. https://doi.org/10.21430/M3OARGGSY0
- Kowarsky M., et al. . 2017. ImmPort. https://doi.org/10.21430/M33PSZ2FHV
- Plunkett J., et al. . 2017. ImmPort. https://doi.org/10.21430/M3AM8G2I2Q
- Zhang G., et al. . 2017. ImmPort. https://doi.org/10.21430/M3F345ZL81
- Stout M. J., et al. . 2018. ImmPort. https://doi.org/10.21430/M3H1U3KJMZ
- Crawford N., et al. . 2018. ImmPort. https://doi.org/10.21430/M3VCNPMD4B
- Rappoport N., et al. . 2018. ImmPort. https://doi.org/10.21430/M37N6PJEQT
References
- Barrett T. et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 41, D991–D995, doi:; DOI: 10.1093/nar/gks1193 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M. & Lash A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikov N. et al. ArrayExpress update--simplifying data submissions. Nucleic Acids Res 43, D1113–D1116, doi:; DOI: 10.1093/nar/gku1057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailman M. D. et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet 39, 1181–1186, doi:; DOI: 10.1038/ng1007-1181 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryka K. A. et al. NCBI's Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res 42, D975–D979, doi:; DOI: 10.1093/nar/gkt1211 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium G. T. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660, doi:; DOI: 10.1126/science.1262110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman R. L. et al. Toward a Shared Vision for Cancer Genomic Data. N Engl J Med 375, 1109–1112, doi:; DOI: 10.1056/NEJMp1607591 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 171, 1437–1452 e1417, doi:; DOI: 10.1016/j.cell.2017.10.049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morken N. H., Kallen K. & Jacobsson B. Outcomes of preterm children according to type of delivery onset: a nationwide population-based study. Paediatric and perinatal epidemiology 21, 458–464, doi:; DOI: 10.1111/j.1365-3016.2007.00823.x (2007). [DOI] [PubMed] [Google Scholar]
- Schaaf J. M., Mol B. W., Abu-Hanna A. & Ravelli A. C. Ethnic disparities in the risk of adverse neonatal outcome after spontaneous preterm birth. Acta obstetricia et gynecologica Scandinavica 91, 1402–1408, doi:; DOI: 10.1111/aogs.12013 (2012). [DOI] [PubMed] [Google Scholar]
- Bastek J. A., Srinivas S. K., Sammel M. D. & Elovitz M. A. Do neonatal outcomes differ depending on the cause of preterm birth? A comparison between spontaneous birth and iatrogenic delivery for preeclampsia. American journal of perinatology 27, 163–169, doi:; DOI: 10.1055/s-0029-1234036 (2010). [DOI] [PubMed] [Google Scholar]
- Green N. S. et al. Research agenda for preterm birth: recommendations from the March of Dimes. American journal of obstetrics and gynecology 193, 626–635, doi:; DOI: 10.1016/j.ajog.2005.02.106 (2005). [DOI] [PubMed] [Google Scholar]
- Ferrero D. M. et al. Cross-Country Individual Participant Analysis of 4.1 Million Singleton Births in 5 Countries with Very High Human Development Index Confirms Known Associations but Provides No Biologic Explanation for 2/3 of All Preterm Births. PLoS One 11, e0162506, doi:; DOI: 10.1371/journal.pone.0162506 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. H. et al. Preventing preterm births: analysis of trends and potential reductions with interventions in 39 countries with very high human development index. Lancet 381, 223–234, doi:; DOI: 10.1016/S0140-6736(12)61856-X (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macones G. A. et al. A polymorphism in the promoter region of TNF and bacterial vaginosis: preliminary evidence of gene-environment interaction in the etiology of spontaneous preterm birth. American journal of obstetrics and gynecology 190, 1504–1508, discussion 1503A, doi:; DOI: 10.1016/j.ajog.2004.01.001 (2004). [DOI] [PubMed] [Google Scholar]
- Roberts A. K. et al. Association of polymorphism within the promoter of the tumor necrosis factor alpha gene with increased risk of preterm premature rupture of the fetal membranes. American journal of obstetrics and gynecology 180, 1297–1302 (1999). [DOI] [PubMed] [Google Scholar]
- Annells M. F. et al. Interleukins-1, -4, -6, -10, tumor necrosis factor, transforming growth factor-beta, FAS, and mannose-binding protein C gene polymorphisms in Australian women: Risk of preterm birth. American journal of obstetrics and gynecology 191, 2056–2067, doi:; DOI: 10.1016/j.ajog.2004.04.021 (2004). [DOI] [PubMed] [Google Scholar]
- Engel S. A. et al. Risk of spontaneous preterm birth is associated with common proinflammatory cytokine polymorphisms. Epidemiology 16, 469–477 (2005). [DOI] [PubMed] [Google Scholar]
- Kalish R. B., Vardhana S., Gupta M., Perni S. C. & Witkin S. S. Interleukin-4 and -10 gene polymorphisms and spontaneous preterm birth in multifetal gestations. American journal of obstetrics and gynecology 190, 702–706, doi:; DOI: 10.1016/j.ajog.2003.09.066 (2004). [DOI] [PubMed] [Google Scholar]
- Wise P. H et al. Risky Business: Meeting the Structural Needs of Transdisciplinary Science. J Pediatr 191, 255–258, doi:; DOI: 10.1016/j.jpeds.2017.08.072 (2017). [DOI] [PubMed] [Google Scholar]
- Eidem H. R., McGary K. L., Capra J. A., Abbot P. & Rokas A. The transformative potential of an integrative approach to pregnancy. Placenta 57, 204–215, doi:; DOI: 10.1016/j.placenta.2017.07.010 (2017). [DOI] [PubMed] [Google Scholar]
- Uzun A et al. dbPTB: a database for preterm birth. Database (Oxford) 2012, bar069, doi:; DOI: 10.1093/database/bar069 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. et al. GEneSTATION 1.0: a synthetic resource of diverse evolutionary and functional genomic data for studying the evolution of pregnancy-associated tissues and phenotypes. Nucleic Acids Res 44, D908–D916, doi:; DOI: 10.1093/nar/gkv1137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S. et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res 58, 234–239, doi:; DOI: 10.1007/s12026-014-8516-1 (2014). [DOI] [PubMed] [Google Scholar]
- Bhattacharya S. et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci Data 5, 180015, doi:; DOI: 10.1038/sdata.2018.15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiulio D. B. et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci USA 112, 11060–11065, doi:; DOI: 10.1073/pnas.1502875112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudilliere B. et al. Implementing Mass Cytometry at the Bedside to Study the Immunological Basis of Human Diseases: Distinctive Immune Features in Patients with a History of Term or Preterm Birth. Cytometry A 87, 817–829, doi:; DOI: 10.1002/cyto.a.22720 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauder A. P. et al. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 4, 29, doi:; DOI: 10.1186/s40168-016-0172-3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman W. E. t. et al. Comprehensive RNA profiling of villous trophoblast and decidua basalis in pregnancies complicated by preterm birth following intra-amniotic infection. Placenta 44, 23–33, doi:; DOI: 10.1016/j.placenta.2016.05.010 (2016). [DOI] [PubMed] [Google Scholar]
- Callahan B. J. et al. Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women. Proc Natl Acad Sci U S A 114, 9966–9971, doi:; DOI: 10.1073/pnas.1705899114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghaeepour N. et al. An immune clock of human pregnancy. Sci Immunol 2, doi:; DOI: 10.1126/sciimmunol.aan2946 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W. et al. Simultaneously Monitoring Immune Response and Microbial Infections during Pregnancy through Plasma cfRNA Sequencing. Clin Chem 63, 1695–1704, doi:; DOI: 10.1373/clinchem.2017.273888 (2017). [DOI] [PubMed] [Google Scholar]
- Kowarsky M. et al. Numerous uncharacterized and highly divergent microbes which colonize humans are revealed by circulating cell-free DNA. Proc Natl Acad Sci U S A 114, 9623–9628, doi:; DOI: 10.1073/pnas.1707009114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plunkett J. et al. An evolutionary genomic approach to identify genes involved in human birth timing. PLoS Genet 7, e1001365, doi:; DOI: 10.1371/journal.pgen.1001365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G. et al. Genetic Associations with Gestational Duration and Spontaneous Preterm Birth. N Engl J Med 377, 1156–1167, doi:; DOI: 10.1056/NEJMoa1612665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout M. J. et al. Early pregnancy vaginal microbiome trends and preterm birth. Am J Obstet Gynecol 217, 356 e351–356 e318, doi:; DOI: 10.1016/j.ajog.2017.05.030 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford N. et al. Divergent Patterns of Mitochondrial and Nuclear Ancestry Are Associated with the Risk for Preterm Birth. J Pediatr, doi:; DOI: 10.1016/j.jpeds.2017.10.052 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappoport N. et al. A genome-wide association study identifies only two ancestry specific variants associated with spontaneous preterm birth. Sci Rep 8, 226, doi:; DOI: 10.1038/s41598-017-18246-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D. et al. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat Commun 8, 1077, doi:; DOI: 10.1038/s41467-017-01027-z (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D., Sirota M. & Butte A. J. Systematic pan-cancer analysis of tumour purity. Nat Commun 6, 8971, doi:; DOI: 10.1038/ncomms9971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley S. C., Sirota M., Chen R., Butte A. J. & Altman R. B. Constraints on Biological Mechanism from Disease Comorbidity Using Electronic Medical Records and Database of Genetic Variants. PLoS Comput Biol 12, e1004885, doi:; DOI: 10.1371/journal.pcbi.1004885 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J. T. et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med 3, 96ra76, doi:; DOI: 10.1126/scitranslmed.3002648 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosti I., Jain N., Aran D., Butte A. J. & Sirota M. Cross-tissue Analysis of Gene and Protein Expression in Normal and Cancer Tissues. Sci Rep 6, 24799, doi:; DOI: 10.1038/srep24799 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirota M. et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med 3, 96ra77, doi:; DOI: 10.1126/scitranslmed.3001318 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirota M., Schaub M. A., Batzoglou S., Robinson W. H. & Butte A. J. Autoimmune disease classification by inverse association with SNP alleles. PLoS Genet 5, e1000792, doi:; DOI: 10.1371/journal.pgen.1000792 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora B. et al. Meta-Analysis of Maternal and Fetal Transcriptomic Data Elucidates the Role of Adaptive and Innate Immunity in Preterm Birth. Front Immunol 9, 993, doi:; DOI: 10.3389/fimmu.2018.00993 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- DiGiulio D. B., et al. . 2016. ImmPort. https://doi.org/10.21430/M3D491LGDT
- Gaudilliere B., et al. . 2015. ImmPort. https://doi.org/10.21430/M3D8CS7ILY
- Lauder A. P., et al. . 2017. ImmPort. https://doi.org/10.21430/M3PZM1ERD2
- Ackerman W. E., et al. . 2017. ImmPort. https://doi.org/10.21430/M34I5YT3K9
- Callahan B. J., et al. . 2017. ImmPort. https://doi.org/10.21430/M37W3869AH
- Aghaeepour N., et al. . 2017. ImmPort. https://doi.org/10.21430/M3OV4WX72N
- Pan W., et al. . 2017. ImmPort. https://doi.org/10.21430/M3OARGGSY0
- Kowarsky M., et al. . 2017. ImmPort. https://doi.org/10.21430/M33PSZ2FHV
- Plunkett J., et al. . 2017. ImmPort. https://doi.org/10.21430/M3AM8G2I2Q
- Zhang G., et al. . 2017. ImmPort. https://doi.org/10.21430/M3F345ZL81
- Stout M. J., et al. . 2018. ImmPort. https://doi.org/10.21430/M3H1U3KJMZ
- Crawford N., et al. . 2018. ImmPort. https://doi.org/10.21430/M3VCNPMD4B
- Rappoport N., et al. . 2018. ImmPort. https://doi.org/10.21430/M37N6PJEQT