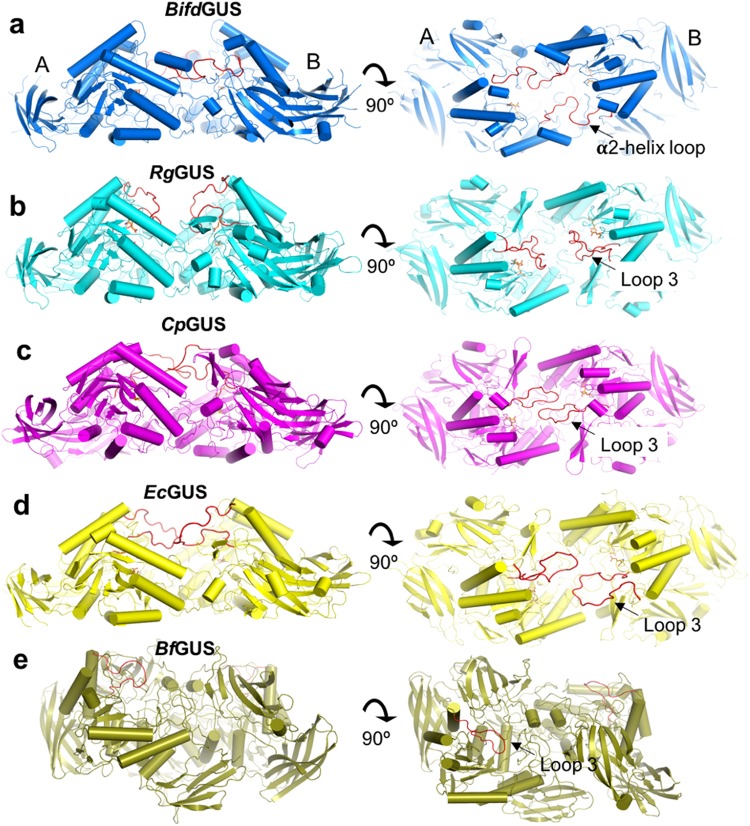
Figure 3.
Comparison of the dimeric structures of the five gut bacterial GUSs. Top and side views of the dimers of the tetrameric (a) BifdGUS, (b) RgGUS, (c) CpGUS (d) EcGUS and (e) BfGUS. The highlighted loops (red) are located either at the periphery of the TIM barrel (loop 3) or at the jelly-roll domain (α2-helix loop). (a) Each α2-helix loop in BifdGUS stretches to the ABS of the adjacent subunit. Each loop 3 of CpGUS (c) and EcGUS (d) exists in an open form and extends from its own subunit to the ABS of the adjacent subunit. Conversely, both loops 3 are not in contact with the adjacent subunit in RgGUS (b). The dimeric interface of BfGUS (e) is oriented differently than in the other GUSs as the active site of the two monomers do not face each other. See also Supplementary Fig. S2. Structural representations were prepared with the use of the Pymol Molecular Graphics System (version 1.8.4.0 Schrodinger).