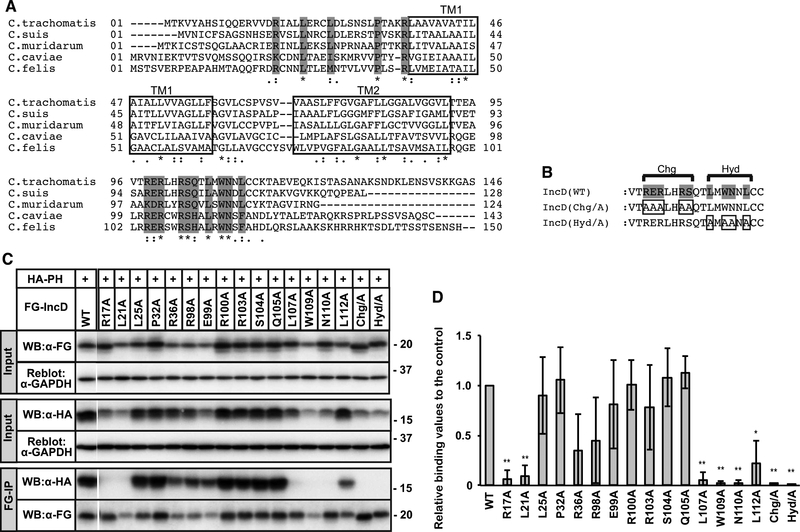
Figure 2. Identification of IncD residues necessary for binding to the CERT PH domain.
(A) Alignment of amino acid sequence of IncD from the indicated Chlamydia species. The predicted TM ™ regions are indicated as white rectangle. Invariant amino acids are shaded grey. (B) The C-terminal amino acids that form the conserved Charged (Chg) and Hydrophobic (Hyd) clusters are indicated. Invariant residues are shaded grey and the residues that were mutated are boxed. (C) HeLa cells were co-transfected with pcDNA4/TO encoding the indicated derivatives of FG-IncD and with pcDNA3.1 neo (+) encoding HA-CERT PH. Samples were prepared as described in Fig. 1. The cell extracts were analysed by Western blotting (Input). The amount loaded as “Input” is 0.6% of the amount used for immunoprecipitation when Western blotting with HA antibody and 2.0% when Western blotting by FLAG antibody. Immunoprecipitated fractions with anti-FLAG antibody (FG-IP) from the cell extracts were analyzed by Western blotting with anti-HA antibody. The membranes were reblotted with an anti-GAPDH antibody. A portion of the gel (between WT and R17A) was spliced out. (D) For the Input fraction and the FG-IP fraction, each band in the image of Western blotting with anti-HA antibody was quantified. The Quantitative values of FG-IP fractions were divided by that of input fractions to correct expression levels and the values are shown as relative values to the control fraction (FG-IncD WT). See Supplementary Material for detail. The data shown are the means ± S.D. from three experiments. *p<0.05 **p<0.01