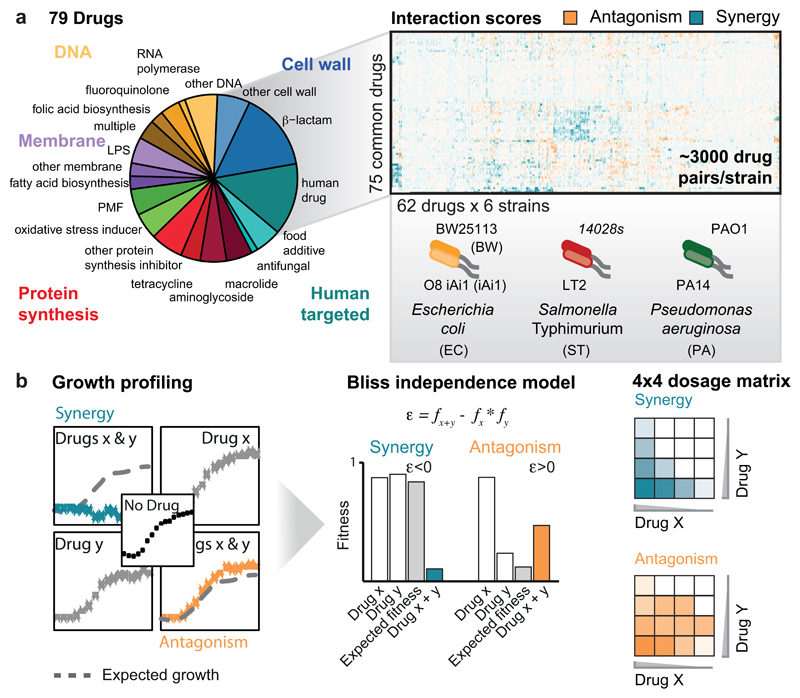
Extended Data Figure 1. High-throughput profiling of pairwise drug combinations in Gram-negative bacteria.
a) Drug and species selection for screen. The 79 drugs used in the combinatorial screen are grouped to categories (Supplementary Table 1). Antibacterials are grouped by target with the exception of antibiotic classes for which enough representatives were screened (>2) to form a separate category: β-lactams, macrolides, tetracyclines, fluoroquinolones and aminoglycosides. Classification of human-targeted drugs and food additives is not further refined, because the MoA is unclear for most. A subset of 62 arrayed drugs was profiled against the complete set of 79 drugs in 6 strains. Strains are color coded according to species. Strain colors and abbreviations are used in all main and ED figures. b) Quantification of drug-drug interactions. Growth was profiled by measuring optical density (OD595nm) over time in the presence of no, single and both drugs. Interactions were defined according to Bliss independence. Significantly lower or higher fitness than expectation (fx*fy) indicates synergy or antagonism, respectively. Synergy and antagonism were assessed by growth in 4x4 checkerboards (Methods).