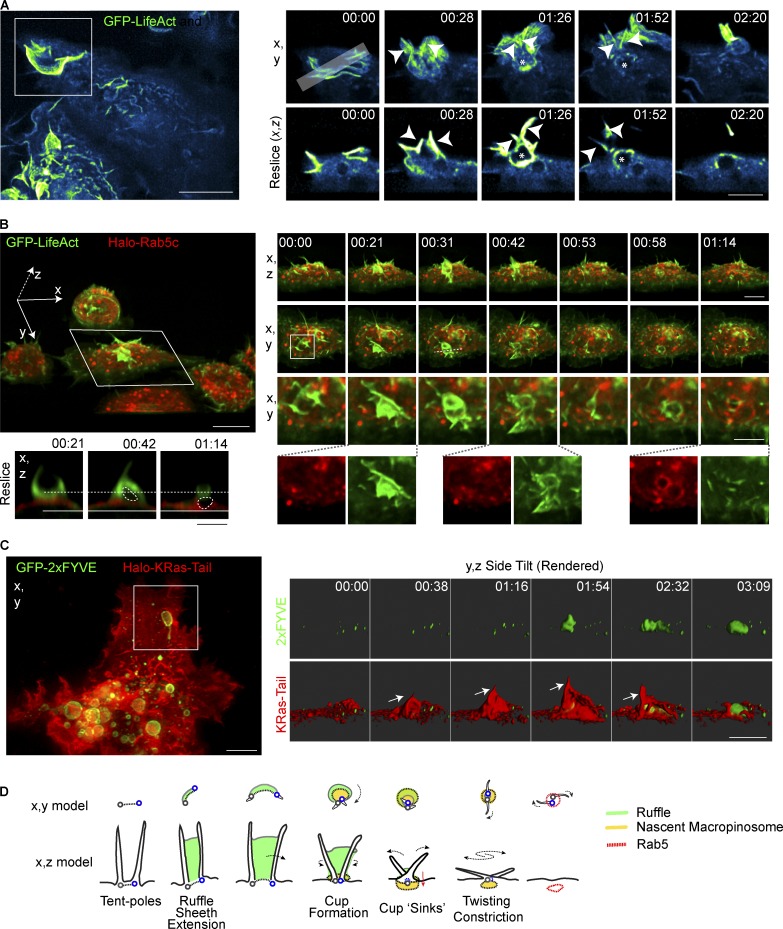
Figure 3.
Tent pole ruffles collapse to form macropinosomes. (A) Macrophages stably expressing GFP-LifeAct. Cells were treated with 100 ng/ml LPS and imaged using LLSM. GFP-LifeAct is represented using the Cyan Hot LUT from ImageJ. Key frames from Video 6. MIP (x,y) and an x,z resliced through the ruffling event is indicated by the gray bar in the top left inset frame. Arrowheads indicate the position of tent poles. Asterisks denotes the macropinosomal void. (B) Macrophages stably expressing GFP-LifeAct and Halo-Rab5c imaged in medium containing LPS. Cells were imaged using LLSM and Halo-tagged proteins were labeled with 10nM JF549. Left large panel shows whole FOV tilted on the x axis from Videos 6 and 7. Small panels show x,z and x,y of boxed cell with second inset box showing one tent pole ruffling event. Reslice of key frames shows the void of the presumptive macropinosome indicated on the merged image with the dotted circle. Separation of the void from the plasma membrane cannot be verified in these images. (C) LLSM of RAW 264.7 macrophages transiently expressing GFP-2×FYVE and Halo-KRas-Tail. Cells were stained with 10 nM JF549 and imaged in media containing LPS. Inset frames were rendered using Imaris and visualized on an x,z side-tilt. Arrow indicates tent pole position in ruffle. (D) Model of macropinosome formation incorporating tent pole crossover and twisting during the constriction phase. Green indicates the ruffle veil, the nascent macropinosome in yellow. The base of each pole is marked as a gray and blue circle. Bars: 10 µm (main panels); 5 µm (inset panels). Time stamps, min:s.