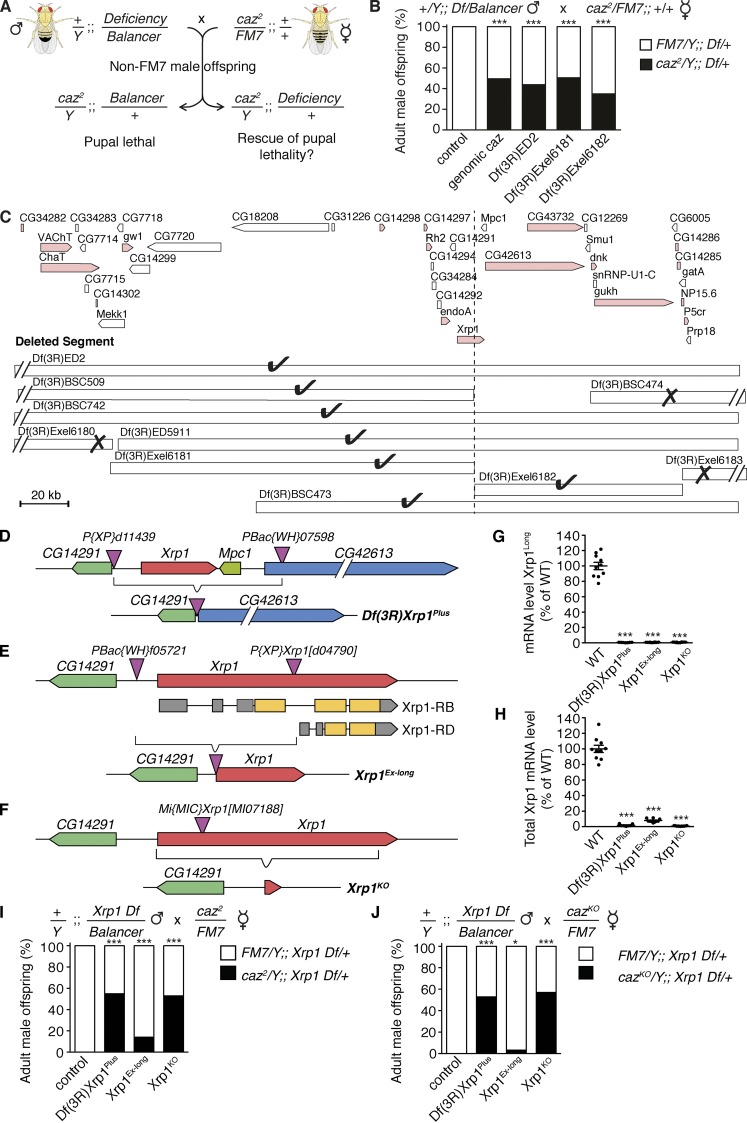
Figure 1. .
Heterozygosity for Xrp1 rescues caz mutant pupal lethality. (A) Screening strategy to identify chromosomal deficiencies that rescue caz2 pupal lethality. (B) Frequency of adult male offspring from the indicated cross that is heterozygous for a genomic caz transgene or the indicated deficiencies. n > 128 per genotype. ***, P < 0.0001; χ2 test. (C) Genomic region uncovered by Df(3R)ED2. Pink indicates genes in the plus orientation; white indicates genes in the minus orientation. The different smaller deficiencies within this region which were tested for rescue of caz2 pupal lethality are shown. Check marks indicate deficiencies that rescue; X marks indicate deficiencies that do not rescue. (D–F) Xrp1 genomic locus showing the insertion sites of the transposable elements used to generate Xrp1 mutant alleles. In the Df(3R)Xrp1Plus allele (D), Xrp1, Mpc1, and the 5′ end of CG42613 are deleted. In the Xrp1Ex-long allele (E), the 5′ half of Xrp1 is deleted, predicted to abolish expression of the Xrp1Long isoform. The Xrp1Short isoform, encoded by Xrp1-RD, may still be expressed. In the Xrp1KO allele (F), the Xrp1 coding region is precisely deleted. (G and H) Xrp1 transcript levels in Xrp1 mutant lines relative to WT controls (100%) determined by qPCR using primers either selectively detecting Xrp1Long transcripts (G) or detecting all Xrp1 transcripts (H). n = 10. ***, P < 0.0001; one-way ANOVA. Mean ± SEM. (I and J) Frequency of adult male offspring from the indicated crosses that is heterozygous for the indicated Xrp1 allele. n > 87 per genotype. *, P < 0.05; ***, P < 0.0001; χ2 test.