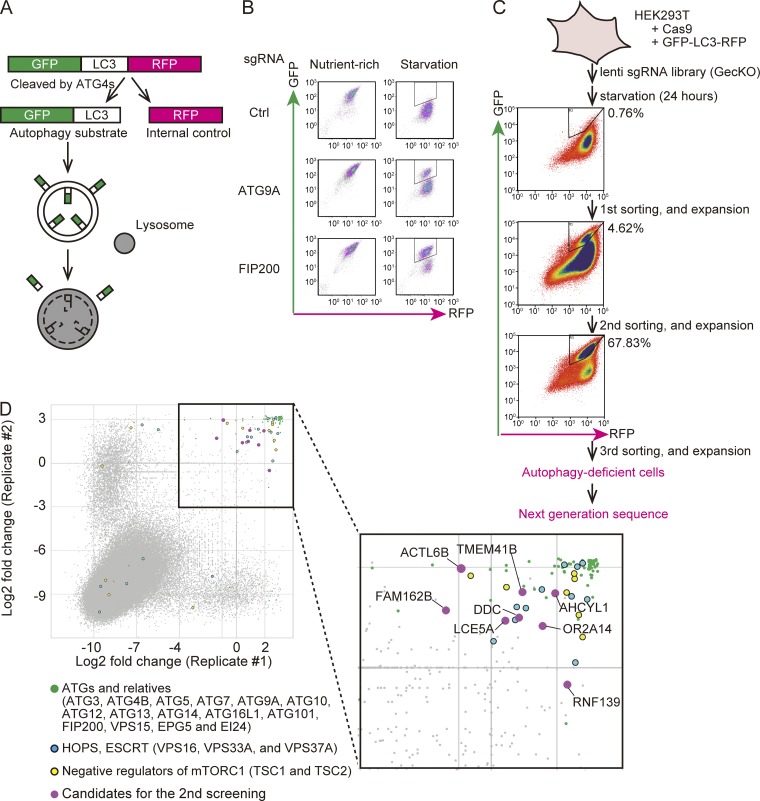
Figure 1.
CRISPR-mediated genome-wide screen using an autophagic flux reporter. (A) Schematic representation of the autophagic flux reporter GFP-LC3-RFP. GFP-LC3-RFP is cleaved by endogenous ATG4 family proteins to yield equimolar amounts of GFP-LC3 (autophagy substrate) and RFP (internal control). Reduction in the GFP:RFP ratio indicates autophagic activity. (B) HEK293T cells expressing Cas9 and GFP-LC3-RFP were transduced with or without sgRNAs targeting ATG9A and FIP200 and selected with puromycin. The GFP and RFP intensities were determined by flow cytometry under nutrient-rich and starvation conditions. The autophagy-deficient population is indicated by the region of interest (ROI). (C) Schematic representation of the CRISPR-mediated genome-wide screen. An sgRNA library (GeCKO) was introduced to HEK293T cells expressing Cas9 and GFP-LC3-RFP. The cell population that did not respond to starvation (indicated by the ROI) was collected by FACS and expanded. After repeating this enrichment process three times, genomic DNA was extracted and subjected to next-generation sequencing. The proportion (%) of the autophagy-deficient population is indicated by the ROI. (D) Scatterplot of the results of two replicates. Data represent log2 (fold change) of read counts of individual sgRNAs before versus after enrichment. Enriched sgRNAs are shown in the separate panel. Canonical ATG genes and known autophagy-related genes (green), genes encoding HOPS and ESCRT components (blue), negative regulators of mTORC1 (yellow), and high-scoring genes not previously linked to autophagy (magenta) are indicated.