Figure 8.
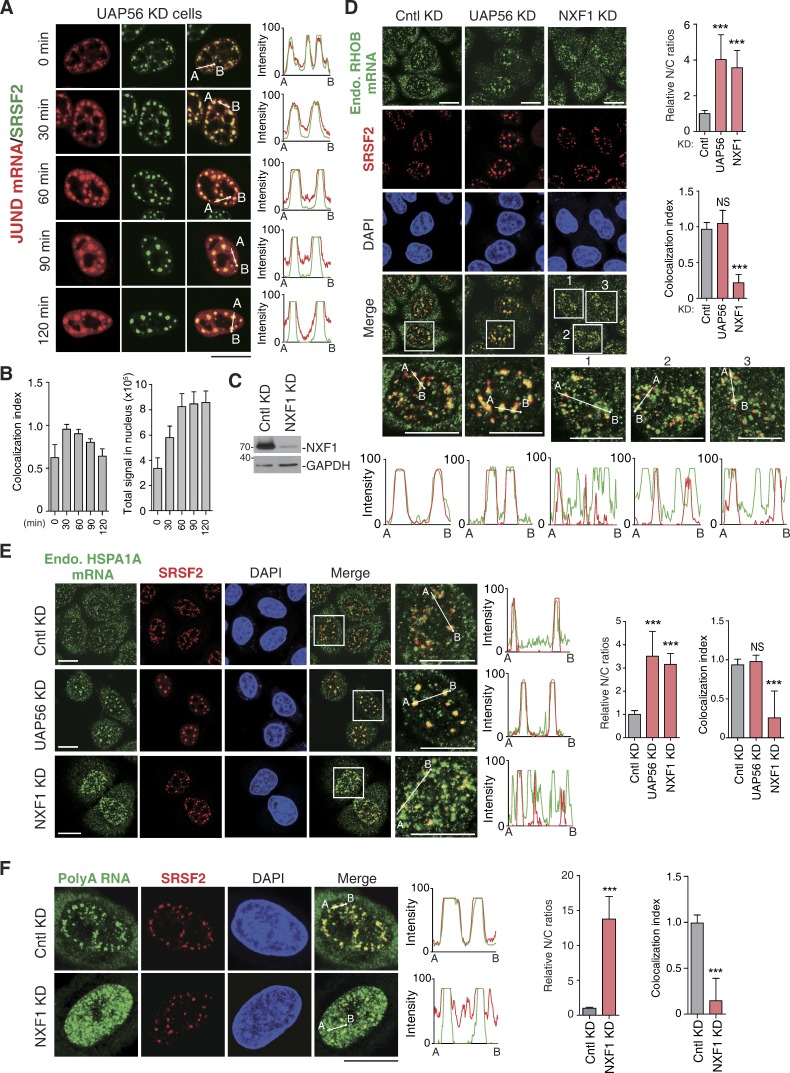
NXF1 depletion results in nuclear mRNA retention both inside and outside of NSs. (A and B) Confocal microscopic images show the JUND mRNA transcribed from the microinjected reporter construct and SRSF2 in HeLa cells treated with UAP56/URH49 siRNA for 48 h. The red and green lines in the graphs show the intensity of the FISH and SRSF2 IF signal, respectively (A). Colocalization indexes of the JUND FISH foci and SRSF2 dots (left) and total FISH signal quantification (right) at each time point are shown in B. (C) Western analysis was used to examine the knockdown (KD) efficiency of NXF1. (D) Confocal microscopic images show the endogenous RHOB mRNA and NSs (SRSF2) in HeLa cells treated with Cntl, UAP56/URH49, or NXF1siRNAs for 48 h. Higher magnification of the boxed regions is shown. The green and red in the line scan graphs demonstrate the intensities of the mRNA and the SRSF2 IF signals, respectively. The bar graphs show the relative N/C ratios (top) and colocalization index (bottom) in each treatment group. Data represent the mean ± SD from three independent experiments; n = 10. (E) Same as in D, except that the endogenous HSPA1A mRNA was examined. (F) Same as in D, except that the bulk polyA RNAs were examined. Bars, 20 µm. Statistical analysis was performed by using the unpaired Student’s t test. ***, P < 0.01. Molecular masses are given in kilodaltons.