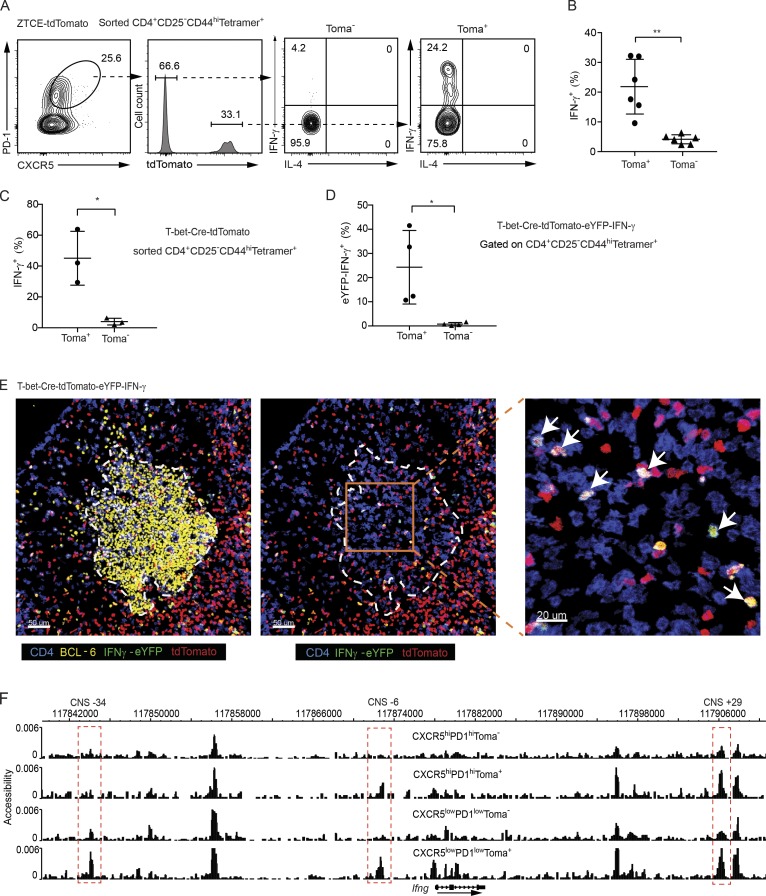
Figure 2.
Ex–T-bet Tfh cells display a partial accessibility at the Ifng locus and have the capability to express IFN-γ. (A–C) Sorted CD4+CD44highCD25−Tetramer+ cells from the immunized TMX-treated ZTCE-tdTomato mice (A and B) or TMX-untreated T-bet-Cre-tdTomato mice (C) were cultured in IL-2–supplemented media overnight. Cells were then stimulated with PMA and ionomycin for 4 h. Cytokine production was assessed by intracellular staining. Percentages of IFN-γ–producing cells were calculated (mean ± SD; in B: **, P < 0.005, n = 6; in C: *, P < 0.05, n = 3). Data are representative of two independent experiments (A–C). (D) Cells from dLN of immunized IFN-γ-eYFP-T-bet-Cre-tdTomato mice were stained with cell surface markers. Percentage of eYFP+ cells among the CD4+CD44highCD25−Tetramer+ tdTomato+ or tdTomato− Tfh (CXCR5highPD-1high) population was calculated (mean ± SD; *, P < 0.05, n = 4). Data are representative of three independent experiments. (E) IFN-γ-eYFP-T-bet-Cre-tdTomato mice were immunized with AS15/CFA. dLN was harvest for imaging analysis. GC (yellow, BCL-6); IFN-γ–expressing ex–T-bet-Tfh cell (shown in white; CD4 shown in blue; tdTomato shown in red; and IFN-γ-eYFP shown in green). Arrows indicate IFN-γ–expressing ex–T-bet-Tfh cells. Data are representative of two independent experiments with two animals in each experiment. (F) CD4+CD44highCD25−Tetramer+-CXCR5highPD-1hightdTomato−, CXCR5highPD-1hightdTomato+, CXCR5lowPD-1lowtdTomato−, and CXCR5lowPD-1lowtdTomato+ cells were sorted from AS15/CFA-immunized T-bet-Cre-tdTomato mice. DHS analysis was performed through scDNase-Seq, and the Ifng gene locus was viewed by the Washington University genome browser. Peaks at the CNS-34, CNS-6 and CNS+29 sites that displayed differential accessibility among the samples are highlighted in red. Cell samples are in biological duplicates.