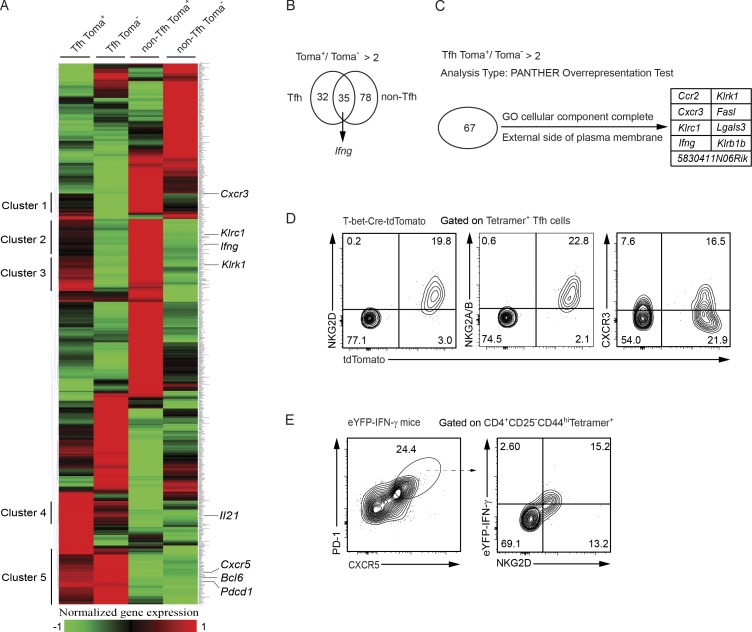
Figure 4.
Unique gene expression profile of the ex–T-bet Tfh cells reveals cell surface markers for identifying this population. (A) RNA-Seq analysis was performed using CD4+CD44highCD25−Tetramer+-CXCR5highPD-1hightdTomato+, CXCR5highPD-1hightdTomato−, CXCR5lowPD-1lowtdTomato+, and CXCR5lowPD-1lowtdTomato− antigen-specific cell populations from dLN of AS15/CFA-immunized T-bet-Cre-tdTomato mice. Differentially expressed genes were clustered. Cell samples are in biological duplicates. (B) Overlapping of the 67 and 113 genes that were expressed at higher levels in tdTomato+ cells compared with tdTomato− counterparts in Tfh and non-Tfh compartment, respectively. (C) tdTomato+ Tfh cell–specific genes (compared with the tdTomato− Tfh cells) that encode cell surface molecules. (D) Antigen-specific Tfh cells from the AS15/CFA-immunized T-bet-Cre-tdTomato mice were stained with anti-NKG2D, anti-NKG2A/B, or anti-CXCR3. Data are representative of three independent experiments. (E) NKG2D and IFN-γ-eYFP expression in antigen-specific Tfh cells from immunized IFN-γ-eYFP mice were measured. Data are representative of two independent experiments with five animals in each experiment.