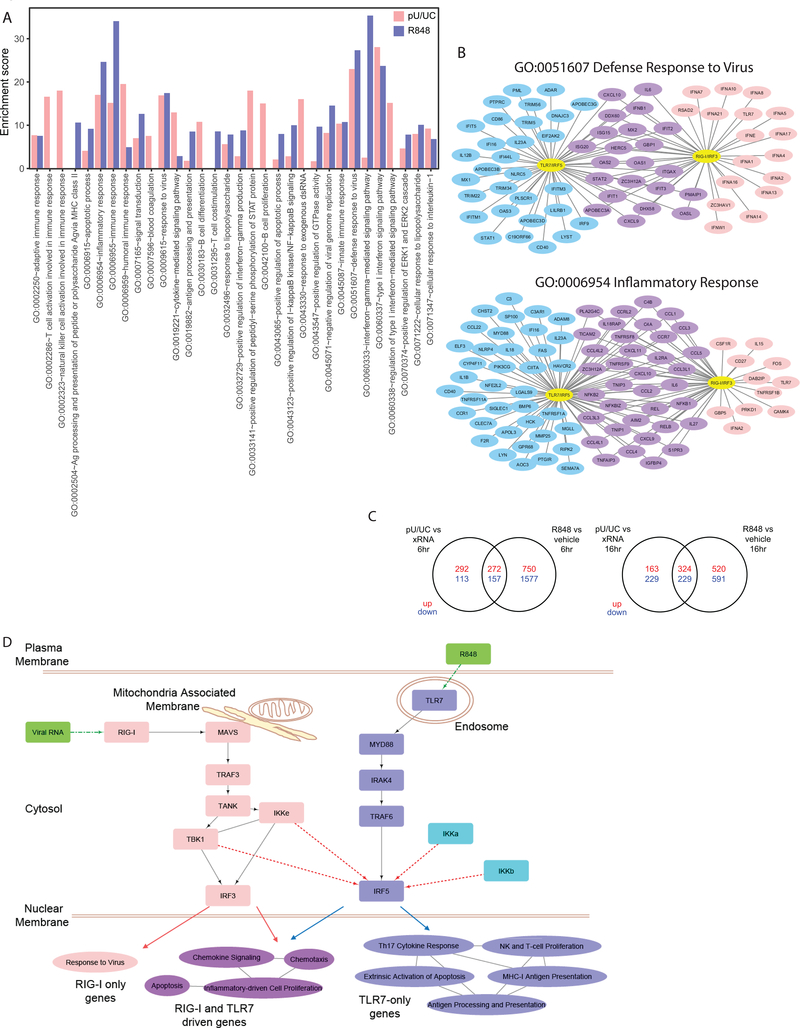
Figure 5. TLR7/IRF5 and RLR/IRF3 regulate genes with distinct functions.
(a) Bar graphs showing the enrichment score of the top 20 most enriched gene ontology (GO) terms in DE genes from R848/vehicle and poly-U/UC/xRNA comparisons. poly-U/UC stimulation is shown in pink, R848 treatment is shown in purple. No bar is shown for categories that are not enriched in the specific comparison. (b) Networks showing genes within GO categories “defense response to virus” (GO:0051607) and “inflammatory response” (GO:0006954) that are DE in R848 treatment (blue), poly-U/UC stimulation (pink), or both (purple). (c) Venn diagrams of upregulated (red) or downregulated (blue) genes in R848/vehicle and pU/UC/xRNA samples at 6 (top) and 16 (bottom) hour time points. (d) Diagram showing RLR and TLR pathway components leading to IRF3 and IRF5 activation using ImmuneDB analysis. Solid lines denote known pathway components. Dotted lines denote potential regulatory relationships demonstrated in our study. Pathway categories enriched in DE gene sets only in R848/vehicle, poly-U/UC/xRNA, or both in RNAseq analysis are displayed on the bottom. Lines between categories denote presence of shared genes in the categories. GO term enrichment induced by poly-U/UC is in pink, those induced by R848 are in blue, those shared by poly-U/UC and R848 stimulation are in purple. RNAseq experiment was performed once with biological triplicates.