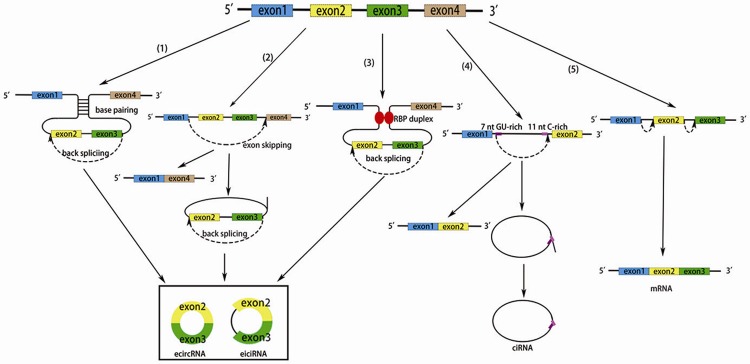
Figure 1.
Possible models of circRNA and mRNA biogenesis. (1) Direct backsplicing. Introns bordering the circularized exons carry out base pairing, which induces the two exons to undergo alternative backsplicing. Introns of the circRNA are removed or retained to form an ecircRNA or eiciRNA. (2) Exon skipping. A downstream exon skips over one or more exons to link an upstream exon, then forms a lariat containing both exons and introns. Introns of the circRNA are removed or retained to form an ecircRNA or eiciRNA. (3) RBP quaking. RBPs bind to recognition elements within introns, then form a bridge between the two flanking intronic sequences, which brings exons in close proximity to undergo backsplicing. Introns of the circRNA are removed or retained to form an ecircRNA or eiciRNA. (4) CiRNA biogenesis. A motif containing an 11 nt C-rich element near the branchpoint and a 7 nt GU-rich element near the 5’ splice escapes the debranching and degradation after the canonical pre-RNA splicing, and forms a ciRNA. (5) Canonical pre-mRNA splicing and mRNA biogenesis.