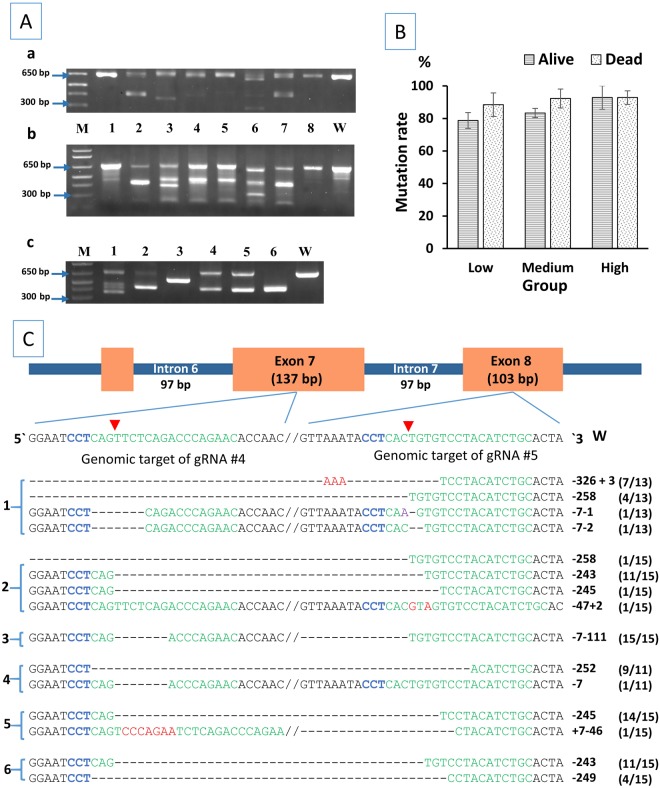
Figure 2.
Analysis of mutagenesis efficiencies of CRISPR/ Cas9 in the rhamnose binding lectin (RBL) gene of channel catfish (Ictalurus punctatus). (A) Identification of mutated RBL gene sequences in channel catfish. One-third of the PCR product from each sample was resolved in a 2% agarose gel to detect the large deletion (a) while two-thirds were digested with Surveyor® enzyme (b) where (a) and (b) came from the same embryo. Samples 1–8 came from embryos microinjected with gRNA/Cas9 protein compared with full-sib wild type control (W). Embryos with a large deletion had shorter band(s) when compared to the wildtype 645 bp band. M indicates 1 kb plus DNA ladder (Invitrogen). (c) PCR products of six microinjected embryo samples that were individually cloned and sequenced where each sample included the DNA from a whole single embryo (sequencing results are illustrated in (C)). (B) Mutation rate ± standard error (SE) of channel catfish dead embryos and alive fingerlings microinjected at the one-cell stage with three dosages of gRNA/Cas9 protein (low dosage: 2.5 ng gRNA/7.5 ng Cas9, medium dosage: 5 ng gRNA/15 ng Cas9 and high dosage: 7.5 ng gRNA/22.5 ng Cas9). (C) CRISPR/Cas9 induced mutations in the RBL gene of channel catfish. Green sequences are the target sites of guide RNAs while the blue sequence (CCT) represents the PAM (protospacer adjacent motif). Red arrows indicate the expected sites of cleavage by Cas9. Samples 1 and 2 were from the low dosage, 3 and 4 from the medium dosage, and 5 and 6 from the high dosage. Dashes and red letters indicate the deletion/insertion of nucleotides, respectively. Numbers on the right side of each sequence indicate the number of nucleotides that have been deleted (−) or inserted (+) e.g. -326-3 means 326 bp deletion and 3 bp insertion. Double slash is present where a DNA sequence between two gRNA targets was omitted for simplicity. Absent double slash means a large deletion where the entire sequence between the two gRNA targets was deleted. Each sequence starting with 5′ and ending with 3′ came from a single reaction representing a single allele. Sequencing reactions with the same type of mutation are represented by a single sequence with a fraction of total e.g. (7/13) means that a certain type of mutation was detected in 7 sequencing reactions out of 13. 2Aa, 2Ab and 2Ac were cropped and the full-length gels are presented in Supplementary Figure S1C, S1D and S1E, respectively.