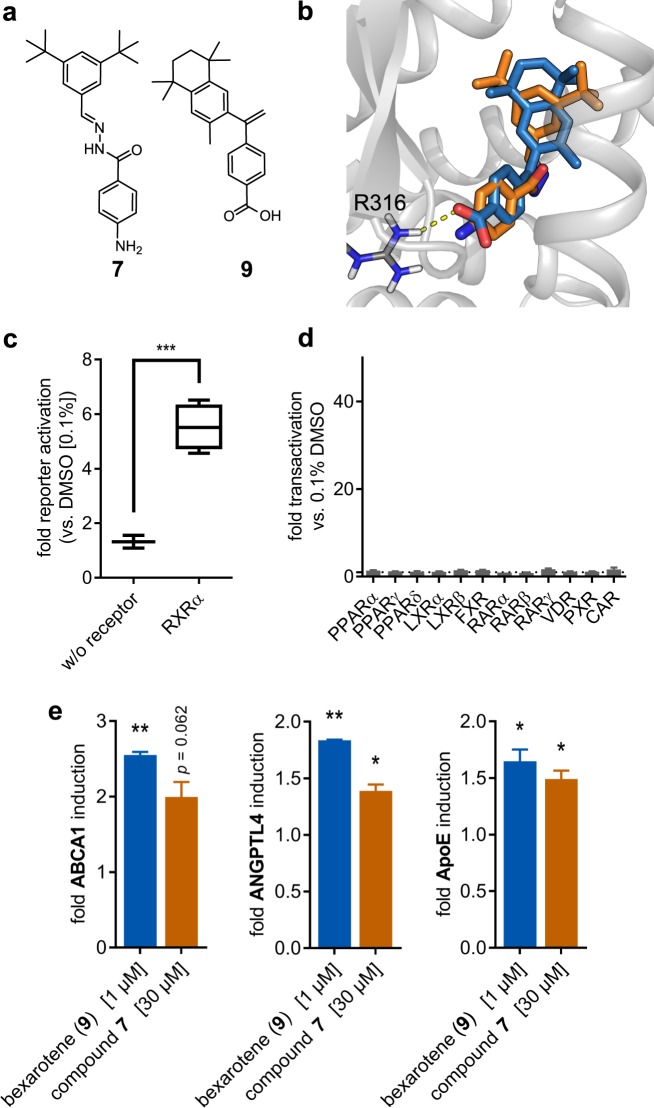
Figure 5.
In silico and in vitro analysis of hit 7. (a) Drug approved RXR agonist bexarotene (9), which was used as the reference for the analysis; (b) Comparison between the predicted binding poses of 7 (orange) and bexarotene (blue) in the ligand binding site of RXRα. The crystal structure of RXRα in complex with the agonist 9cUAB30 and the coactivator peptide GRIP-1 (PDB-ID: 4K4J) was prepared in MOE (v2016.0802)57, following the default protein preparation protocol. Structure energy was minimized using Amber10:EHT force field. For each ligand (i.e., crystalized ligand, bexarotene and hit 7) 60 poses were generated, their energy was minimized using MMFF94x force field within a rigid receptor, and they were ranked by London dG score57; the top 10 poses were refined and scored using GBVI/WSA dG57 and the top-scoring pose was chosen. 7 and bexarotene share a similar binding pose, with 7 missing the interaction with R316 due to its lack of an acidic feature. (c) Control experiment: In absence of a Gal4-RXR hybrid receptor, the Gal4-responsive reporter gene was not transactivated by 7 confirming RXR-mediated activity. (d) RXR ligand 7 is highly selective over twelve related nuclear receptors (peroxisome proliferator-activated receptor [PPARα/γ/δ], liver X receptor [LXRα/β], farnesoid X receptor [FXR], retinoic acid receptor [RARα/β/γ], Vitamin D Receptor [VDR], pregnane X receptor [PXR], constitutive androstane receptor [CAR]). (e) RXR modulator 7 induces RXR regulated genes ATP-binding cassette transporter A1 (ABCA1), angiopoietin like protein 4 (ANGPTL4) and Apolipoprotein E (ApoE) with an efficacy comparable to RXR agonist bexarotene.