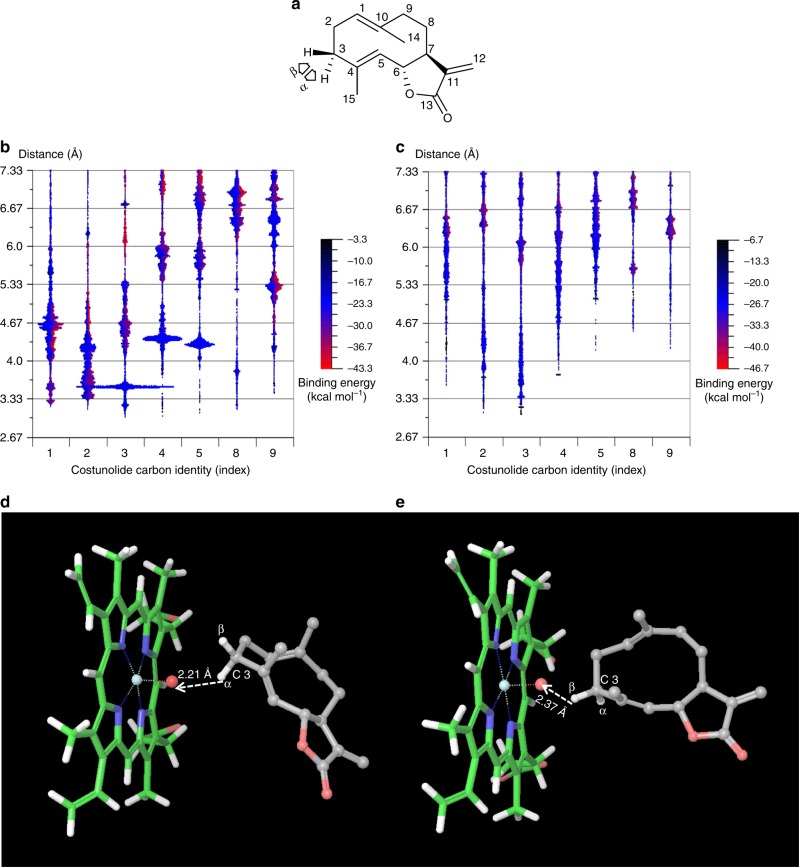
Fig. 3.
In silico docking of costunolide into the active site of the feverfew enzymes, kauniolide synthase and costunolide 3β-hydroxylase. a Structure of costunolide and pro-α and pro-β hydroxylation orientations. b Preferred docking orientations of costunolide as calculated using Protein Energy Landscape Exploration software. Costunolide carbon distance distribution relative to the haem oxyanion in a KLS homology model (constrained model, at 15 Å). c Costunolide carbon distance distribution relative to the haem oxyanion in a costunolide 3β-hydroxylase homology model (constrained model, at 15 Å). In silico docking of costunolide into the active site of kauniolide synthase and costunolide 3β-hydroxylase by Glide. d Preferred docking of costunolide in the active site of kauniolide synthase is in α-orientation. e Preferred docking of costunolide in the active site of costunolide 3β-hydroxylase is in β-orientation. Colour keys of b, c represent the binding energy (kcal mol−1) expressed in negative values