FIGURE 1.
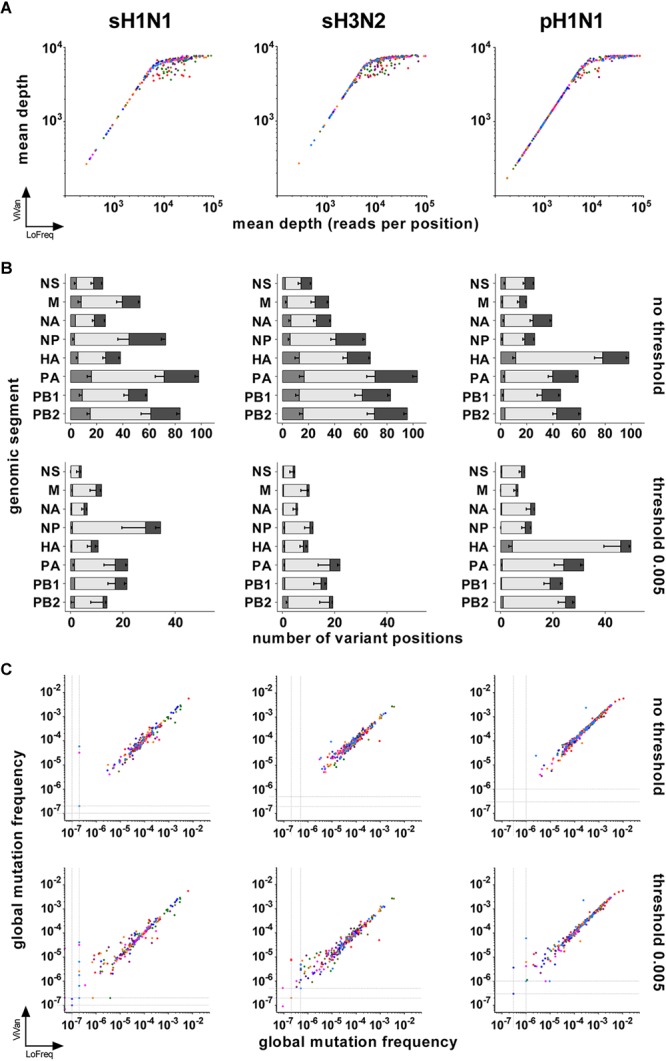
Comparison of LoFreq and ViVAN. (A) Correlation of the mean depths of sequencing (in number of reads per position) obtained by both pipelines. X-axis: LoFreq; Y-axis: ViVAN. Each dot represents one segment of one sample: polymerase sub-unit PB2: olive; polymerase sub-unit PB1: green; polymerase sub-unit PA: purple; hemagglutinin HA: red; nucleoprotein NP: dark blue; neuraminidase NA: pink; matrix and ion channel proteins M: orange; non-structural protein and nuclear export protein NS: light blue. (B) Mean number of variant positions identified by the pipelines, without or with setting a mutation frequency threshold. For each bar, from left to right: gray, specific to LoFreq; light gray, common to LoFreq and ViVAN; dark gray, specific to ViVAN. Error bars represent the Standard Error to the Mean. (C) Correlation of the global mutation frequencies obtained with both pipelines with or without setting a mutation frequency threshold. X-axis: LoFreq; Y-axis: ViVAN. Each dot represents one segment of one sample: PB2: olive; PB1: green; PA: purple; HA: red; NP: dark blue; NA: pink; M: orange; NS: light blue.
