Fig. 1.
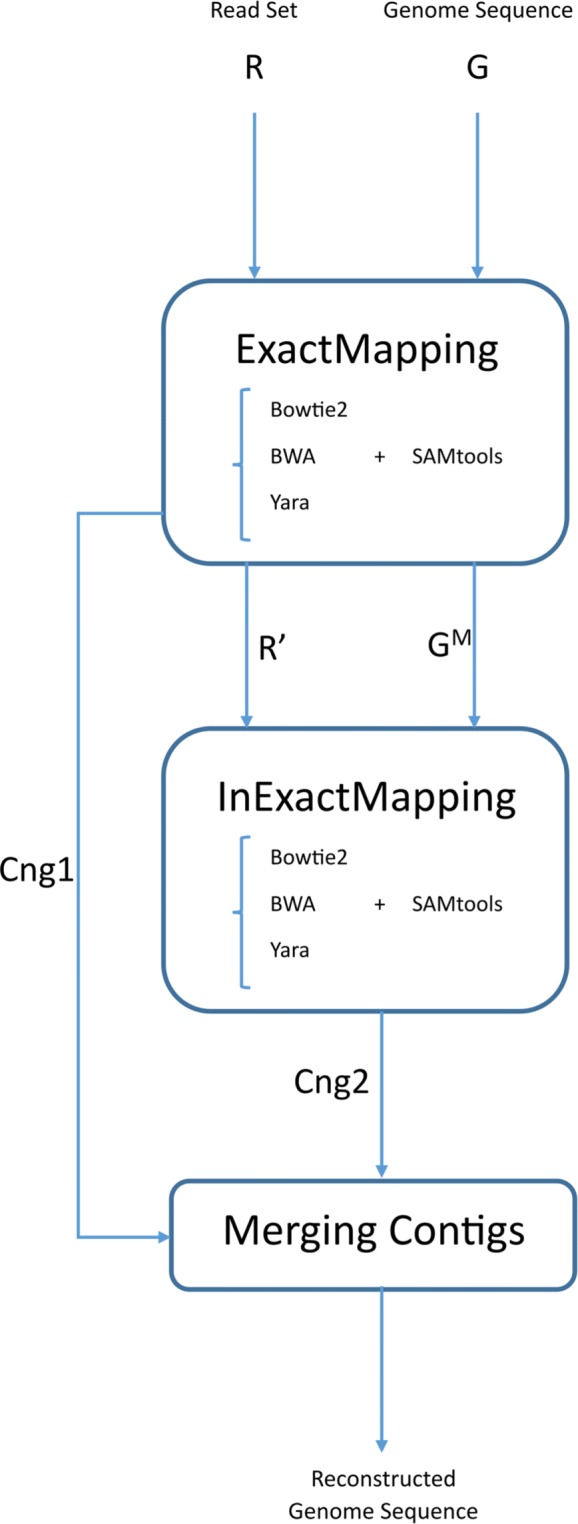
EIM pipeline overview and applied tools. The first step has three outputs: leftover reads (R′), modified remaining parts of the genome sequence (GM) and exact contigs (Cng1). The output of the second step is inexact contig set indicated by Cng2. Currently, EIM can apply one of mappers Bowtie2 [7], BWA [8, 9] and Yara [10] for mapping reads
