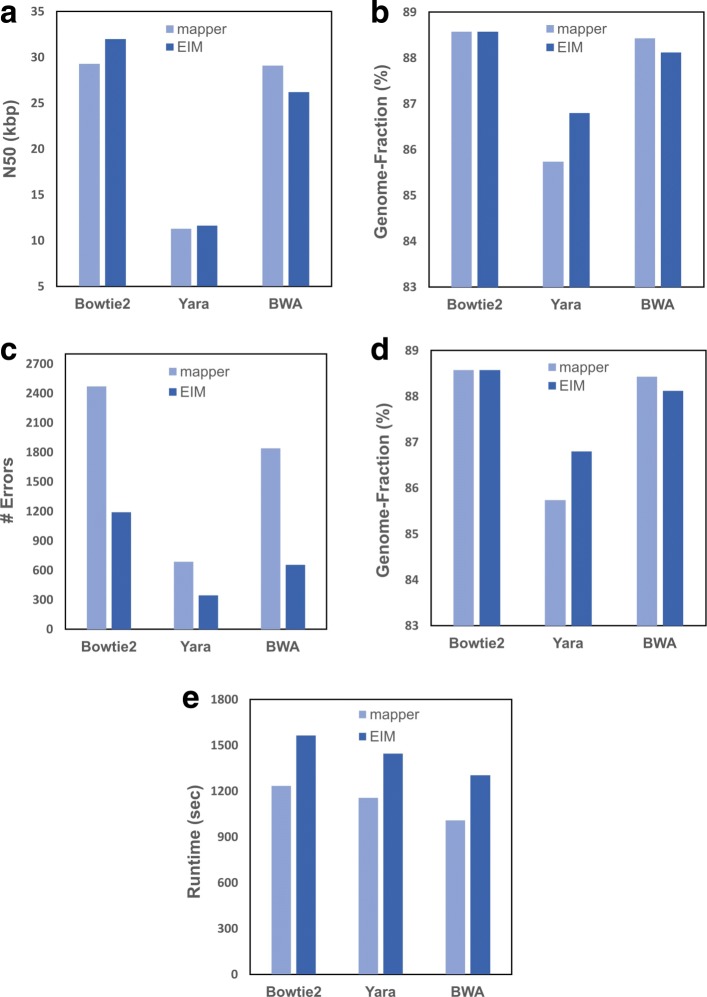
Fig. 2.
The comparison of contigs generated by Bowtie2, Yara and BWA with the respective contigs of EIM on the real read set of E. coli K12. Firstly, the mappers were executed on the read set and the reference, and then the contig sets were generated. Secondly, for each mapper, EIM (v2) was run on the read set and the contig set constructed by the mapper while using it at the second step for mapping. Finally, the contiguity and quality of contigs were computed as a N50 size b Genome-Fraction value c The number of errors d The number of IUPAC codes. In addition, the running time of obtaining contigs was measured and showed in seconds (e)