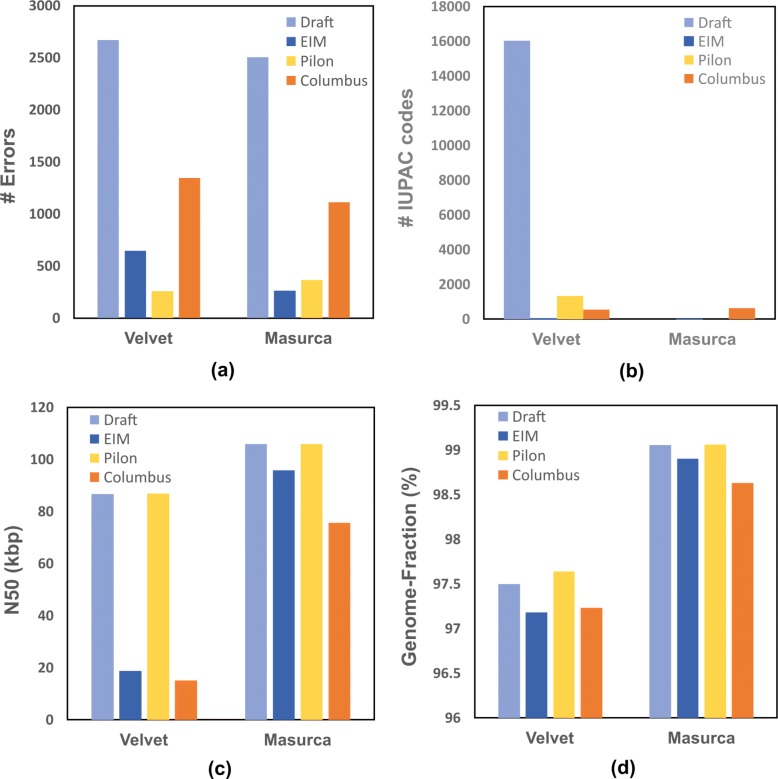
Fig. 3.
The comparison of contigs generated by EIM, Pilon and Columbus on the real read set of E. coli K12.Firstly, two draft genomes were generated by Velvet and MaSuRCA de novo assemblers. Secondly, EIM, Pilon and Columbus were run by each draft and mapped reads on the draft. Finally, the quality and contiguity of contigs were computed as a The number of errors b The number of IUPAC codes c N50 size d Genome-Fraction value