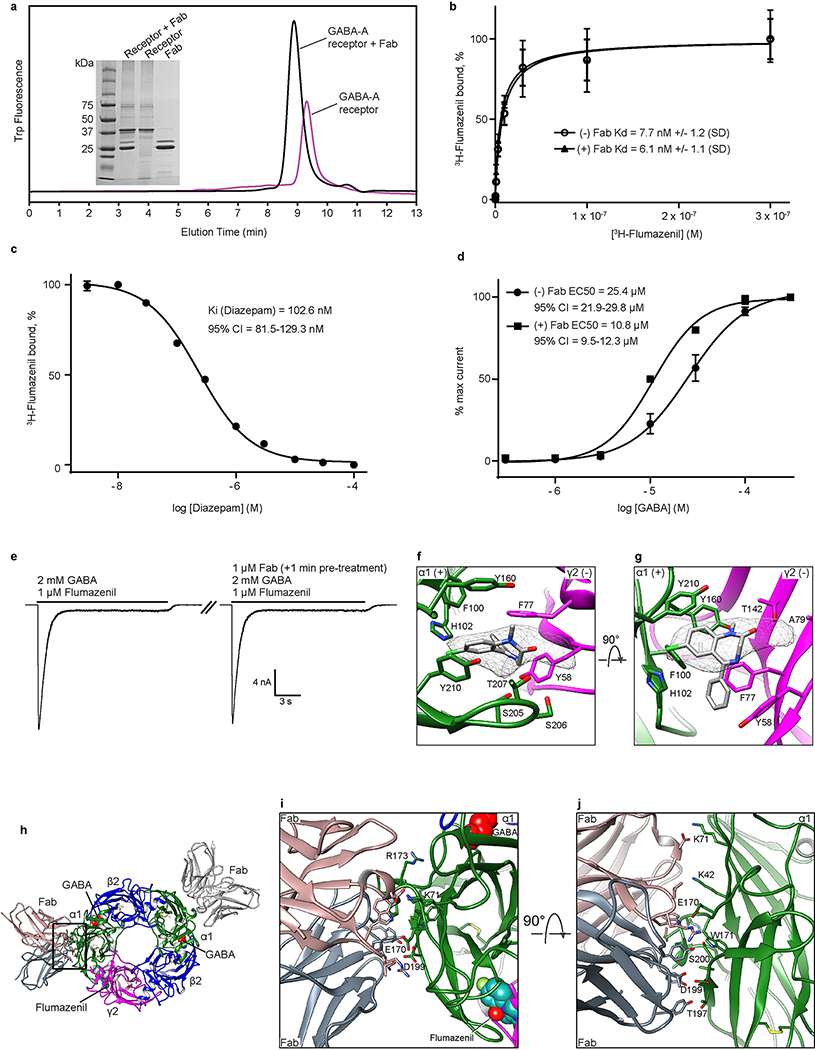
Extended Data Figure 2. Biochemistry and binding assay.
a, FSEC of GABA-A receptor with and without Fab bound and SDS-PAGE analysis of a representative purification (from n>10 purifications). b, Saturation binding assay with [3H]-flumazenil. Single site binding fits for receptor alone and receptor plus Fab both exhibited a Hill slope of ~1 (0.97 and 0.89 respectively). Plotted results are from a representative experiment performed in triplicate. n=3 independent experiments. Data point center is the mean. Error bars are standard deviation, shown for a representative triplicate measurement. c, Competition of 10 nM [3H]-flumazenil with diazepam. Calculated Ki for diazepam assumes a Kd of [3H]-flumazenil of 7.7 nM. n=2 independent experiments in triplicate. Error bars are standard error of the mean (s.d.), shown for a representative triplicate measurement. d, Dose-response experiments in the presence or absence of Fab. HEK cells were transfected with EM constructs and patch-clamped with or without pretreatment of 1 μM Fab for one minute. Hill slopes are 1.7 and 1.4 with and without Fab, respectively. Published values for GABA EC50 range from 6.6 μM – 107 μM71–74. n=3 experiments from different cells. Data point center is the mean. Error bars are standard deviation. e, Whole cell patch clamp recording of long application of EM ligands at concentrations used in EM sample to assess conformational state at equilibrium. The two traces shown are from one continuous recording; in between the two responses, Fab was added to 1 μM for one minute to saturate all receptor sites before second application of GABA and flumazenil (including Fab). n=3 independent experiments. f-g, Docking of diazepam at the benzodiazepine binding site based on superposition of benzodiazepine rings. The phenyl ring of diazepam would orient toward the membrane, possibly forming π-π stacking interactions with Y58 on the complementary subunit. Similar to flumazenil, the halogen of diazepam could interact with H102, suggesting this contact is conserved broadly among benzodiazepines and flumazenil. This orientation is largely consistent with predictions from a modeling and docking study75 and distinct from that suggested by affinity labeling76. In this latter prediction, the diazepam phenyl group orients away from the membrane and would require local reorganization of side chains to avoid atomic clashes. h-j, Structural details of Fab-α1 interaction. Labeled residues are on α subunit. i, Top view. j, Side view.