Figure 1.
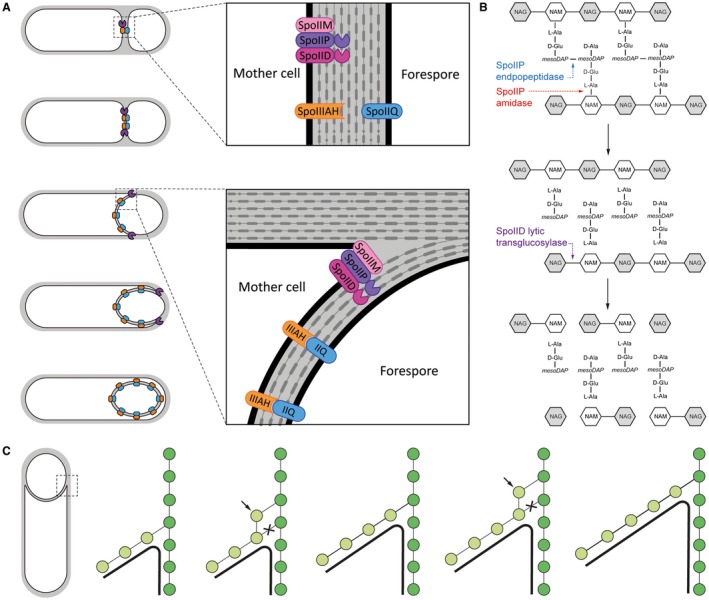
Molecular basis of engulfment. (A) Schematic representation of engulfment, as proposed for B. subtilis. (i) Following asymmetric cell division, components of the SpoIIDMP engulfment machinery (purple) are recruited to the septal midpoint, anchoring themselves within the mother cell membrane. (ii) The complex is then involved in thinning septal peptidoglycan as the machinery relocates to the leading edge of the engulfing membrane, while SpoIIQ (blue) and SpoIIIAH (orange) co‐localize at the mother cell/forespore interface. (iii) During engulfment, the SpoIIDMP complex ‘pulls’ the mother cell membrane around the forespore while being trailed by SpoIIQ and SpoIIIAH which interact within the intermembrane space, preventing membrane retraction. (iv) After completion of engulfment, the SpoIIDMP complex dissociates while the SpoIIQ and SpoIIIAH form a scaffold for assembly of a secretion system that spans the intermembrane space and ‘nurtures’ the spore. Adapted from Sogaard‐Andeersen, 2013. (B) Sequential breakdown of PG by SpoIID and SpoIIP. (i) SpoIIP endopeptidase activity removes peptide crosslinks while its amidase activity removes peptide stems from the glycan chain. (ii) The denuded glycans are then processed by SpoIID’s lytic transglycosylase activity, releasing NAG‐anhydroNAM disaccharides. NAG: N‐Acetylglucosamine; NAM – N‐acetylmuronic acid. (C) Cell cross‐section with glycan strands in the plane perpendicular to the long axis of the cell. One strand from old cell wall (dark green) and one strand from newly synthesized spore‐cell wall (light green) are used as a template for new glycan insertion. Coordination between glycan insertion (arrow) and peptide cross‐link degradation (cross) drives the engulfing membrane (black) forward. Adapted from Ojkic et al., 2016.
