Abstract.
Carrion’s disease is a neglected, vector-borne illness that affects Colombia, Ecuador, and especially Peru. The phlebotomine sand flies Lutzomyia verrucarum and Lutzomyia peruensis are the main illness vectors described, although other species may be implicated in endemic areas such as some northern Peruvian regions, in which Carrion’s disease vector has not been established. The aim of this study was to evaluate the presence of Bartonella bacilliformis DNA in Lutzomyia maranonensis from Cajamarca, northern Peru. This sand fly has not been defined as a vector yet. Centers for Disease Control and Prevention light traps were used to collect adult phlebotomine sand flies from 2007 to 2008 in the Cajamarca department. Female specimens were identified using morphological keys and were grouped into pools of five sand flies, taking into account district and sampling site (intradomicile or peridomicile). DNA was extracted, and then conventional and real-time polymerase chain reaction (RT-PCR) were performed to detect B. bacilliformis and subsequently confirmed by sequencing. A total of 383 specimens of L. maranonensis species were analyzed. Two of 76 pools were positive for B. bacilliformis by sequencing; all positives pools were from Querocotillo district. In addition, Mesorhizobium spp. were identified in two pools of sand flies, which is an α-proteobacteria phylogenetically very close to B. bacilliformis. This study presents molecular evidence that suggests L. maranonensis is naturally infected by B. bacilliformis in the Cajamarca department. Further research should determine if L. maranonensis is a vector and could transmit B. bacilliformis.
INTRODUCTION
Carrion’s disease is a vector-borne disease caused by the bacterium Bartonella bacilliformis and is considered a true neglected illness that not only affects the vulnerable populations in the Andean regions of Peru but has also been reported in Colombia and Ecuador.1,2 Carrion’s disease or bartonellosis is a biphasic illness with acute and chronic phases.1–3 In the acute, so-called Oroya fever, several unspecific symptoms are present, including anemia, jaundice, malaise, or fever, among others,4 which in the absence or delay of adequate treatment may lead to fatality rates as high as 88% due to high bacteremia, transient immunosuppression, and the presence of opportunistic infections.1–3 The distinctive trait of the chronic phase, so-called Peruvian wart, is a series of verrucous lesions due to an uncontrolled endothelial cell proliferation.3 There are around 40–50% of asymptomatic carriers in endemic areas who, in the absence of an animal reservoir, are considered the perpetuators of the disease.5
Peru has endemic areas with approximately 1.7 million at-risk inhabitants.1–3 Moreover, in the last few years, an expansion of this disease toward new areas has been reported.6 Classically, the distribution of the illness has been confined to the inter-Andean valleys, at 500–3,200 m of altitude.1 This distribution is mainly related to the ecology of the sand fly Lutzomyia spp. There are approximately 480 species of sand flies of the genus Lutzomyia, and 149 species have been described in Peru alone.7 Of these, two species have been incriminated as vectors of Carrion’s disease. Lutzomyia verrucarum is the main vector described in the highlands,3,4,8 and Lutzomyia peruensis, the presence of which has not only been mostly described in southern Peru9 but also in specific focalized regions of Inter-Andean valleys of central and northern Peru.10 However, a significant prevalence of Carrion’s disease has also been found in areas where L. verrucarum or L. peruensis is not present, indicating endemic conditions. It is hypothesized that other Lutzomyia species, present in these areas, could be vectors, such as Lutzomyia pescei, Lutzomyia robusta, Lutzomyia maranonensis, or Lutzomyia serrana.11,12 Nonetheless, none of these potential vectors has been confirmed to date. Indeed, vectors of Carrion’s disease need to be identified in the northern department of Cajamarca, where it is ranked third in probable cases reported.13 In Cajamarca, as well as in the neighboring Amazonas department of Peru, L. maranonensis is considered a potential vector of Carrion’s disease. Lutzomyia maranonensis was among the most frequently captured species in Cajamarca and Amazonas.11 Moreover, L. maranonensis presented a higher anthropophilic indoor behavior than other sand fly species. Therefore, L. maranonensis is adapted to peridomiciliary environments and is most frequently captured when using human bait capture.11
These gaps in knowledge are of special concern to understand both the epidemiology and the potential for dissemination of the disease, as well as to define potential disease control actions and its future elimination.4 Thus, the aim of this study was to evaluate the presence of B. bacilliformis DNA in populations of L. maranonensis from Cutervo Province in the Cajamarca department, northern Peru, to define if this sand fly species could be a potential vector of Carrion’s disease.
MATERIAL AND METHODS
Study site.
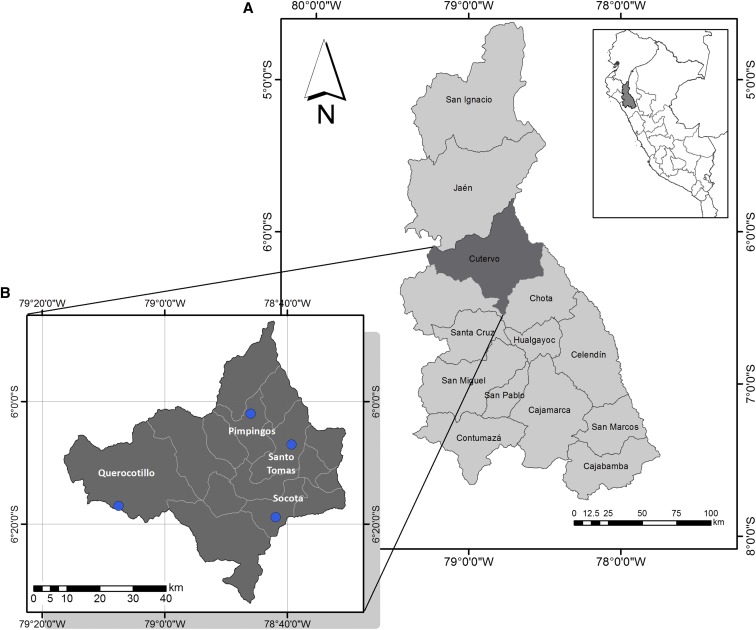
Lutzomyia maranonensis specimens were captured from December 2007 to February 2008 at four sampling sites in Cutervo Province (Cajamarca department, northern Peruvian highlands): Coro Alto (1,871 m), Sucse (1,971 m), El Laurel (1,422 m), and Succha (2,143 m), belonging to the Querocotillo, Socota, Pimpingos, and Santo Tomas districts, respectively (Figure 1). Cutervo is located on the eastern slopes of the Andean mountains. It has a semidry, temperate climate. The average maximum annual temperature is 22°C, with a minimum of 5°C. The rainy season usually starts in November and ends in April.14
Figure 1.
Geographical location of sampling points in the study. (A) Map of Cajamarca department. (B) Map of Cutervo Province. This figure was elaborated using software ArcGIS 10.3 (Environmental Systems Research Institute, Redlands, CA). This figure appears in color at www.ajtmh.org.
Sand fly collection.
Sand flies were captured and stored by Abraham Cáceres of the Entomology Laboratory of Tropical Medicine Institute of Universidad Nacional Mayor de San Marcos. Specimens were captured with CDC light traps installed in a number of intradomicile (Querocotillo, Socota, and Pimpingos) or peridomicile (Santo Tomas) locations. The sand flies were morphologically identified based on measurements of the delta portion of wing, spermatheca, and cibarium under stereomicroscopy.15 Once dissected, pools of up to five female sand flies from each site that had been identified and confirmed as L. maranonensis species were separated, stored in tubes, and sent to the Molecular Laboratory of the Universidad Peruana de Ciencias Aplicadas. Then, the pools were immersed in 70% ethanol and stored at −20°C, until molecular testing.
DNA extraction.
Sand fly pools were crushed, and the DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions except that the final elution volume was 100 μL. The samples were stored at 4°C until used, being thereafter stored at −20°C.
Conventional PCR analysis.
Prokaryotic ribosome RNA primers, targeting the 16S rRNA gene, were used. These primers can detect positive Bartonella spp. samples by amplifying a conserved 438-bp fragment.16 All samples were run in duplicate. The B. bacilliformis collection strain (CIP 57.19, NCTC 12135) was used as the positive control, whereas a PCR reaction without template DNA was used as the negative control.
Real-time PCR analysis.
Previously described RT-PCR targeting the 16-23S intergenic transcribed spacer region of B. bacilliformis was used, according to Dong-Mei et al.17 Briefly, RT-PCR conditions were 95°C for 15 minutes, 50 cycles of 20 seconds at 95°C, 30 seconds at 60°C, and 15 seconds at 72°C. Each reaction contained 5 μL of template DNA and 15 μL of RT-PCR master mix including 1 μL (10 µM of each primer and 1.2 μL (10 µM) of Taqman probe. The controls were the same as conventional PCR analysis.
Sequencing.
Five conventional PCR amplicons with the expected size were recovered QIAquick PCR purification Kit (Qiagen) and sequenced by MACROGEN service (Seoul, Korea).
Statistical analysis.
Descriptive statistics were performed using Stata software (version 14.0; StataCorp LP, College Station, TX). Positive predictive values (PPVs) with 95% confidence intervals (95% CIs) were calculated for conventional PCR tests using sequencing test as the gold standard. The minimal infection rate (MIR) was calculated as described by Paiva et al.18: MIR = no. of positive pools × 100/total no. of insects.
RESULTS
A total of 383 sand flies morphologically identified as L. maranonensis females were analyzed in 76 pools of up to five individuals each. Most sand flies were from Querocotillo (53.5%), followed by Pimpingos (29.0%), Santo Tomas (9.1%), and Socota (8.4%). Twelve pools (15.8%) were positive, by either conventional PCR (8, 10.5%) or RT-PCR (7, 9.2%), respectively. Three pools were positive by both conventional PCR and RT-PCR, and two of them were also positive for B. bacilliformis when sequenced (2.6%, 95% CI: 0.3–9.2%) (Table 1). The sequences showed a 100% identity with the 16S rRNA gene of B. bacilliformis (GenBank accession number CP014012.2). In addition, five pools were positive by conventional PCR but negative by RT-PCR, and the sequences of two pools detected the presence of Mesorhizobium spp. (GenBank accession number KY653035.1). This suggests that false-positive results are possible by PCR (Table 2).
Table 1.
Sand fly sample characteristics
| District | Habitats | Sand flies | Pools | Confirmed positive pools (minimal infection rate) | |||
|---|---|---|---|---|---|---|---|
| # | Positive for Bartonella bacilliformis | ||||||
| N (%) | Conventional PCR | RT-PCR | Sequencing | ||||
| Querocotillo | Intradomicile | 205 (53.5) | 41 | 5 | 6 | 2 | 2 (1.0) |
| Pimpingos | Intradomicile | 111 (29.0) | 22 | 1 | 1 | 0 | 0 (0.0) |
| Santo Tomas | Peridomicile | 35 (9.1) | 7 | 2 | 0 | 0 | 0 (0.0) |
| Socota | Intradomicile | 32 (8.4) | 6 | 0 | 0 | 0 | 0 (0.0) |
| Total | 383 | 76 | 8 | 7 | 2 | 2 (0.5) | |
Table 2.
Positive pools for any of the methodologies used
| District | Pools | Conventional PCR | RT-PCR | Sequencing |
|---|---|---|---|---|
| Querocotillo | I | − | + | ND |
| II | − | + | ND | |
| III | + | + | Bartonella bacilliformis | |
| IV | − | + | ND | |
| V | + | + | B. bacilliformis | |
| VI | + | + | ND | |
| VII | + | − | ND | |
| VIII | + | − | ND | |
| Pimpingos | IX | + | − | Mesorhizobium spp. |
| X | − | + | ND | |
| Santo Tomas | XI | + | − | Mesorhizobium spp. |
| XII | + | − | ND |
ND = not done.
Among eight pools positive by conventional PCR, only two were confirmed by sequencing as positive for B. bacilliformis, resulting in a PPV of 25% (95% CI: 3.2–65.1%). In other words, the false-positive rate of the conventional PCR assay used in this study was 34.9%. All positive pools were from Querocotillo district, where the individual MIR was 0.98 pools positive per 100 sand flies (95% CI: 0.94–0.99) and the general MIR of this study was 0.5 (95% CI: 0.5–0.6) (Table 1).
DISCUSSION
We observed molecular evidence of B. bacilliformis DNA in L. maranonensis in the Cajamarca department, Peru. Two of three sand fly pools were positive by highly specific sequencing and RT-PCR assays. Five conventional PCR+ pools were not sequenced for insufficient DNA quantity/quality. Despite this limitation, the B. bacilliformis DNA evidence suggests that L. maranonensis is a potential vector of B. bacilliformis and further research should determine if it is a vector and can transmit B. bacilliformis. Our findings are consistent with the circulation of B. bacilliformis in this region in the absence of other known vectors.11 The role of L. maranonensis as a potential vector is also consistent with the historical circulation of B. bacilliformis in the areas of northern Peru with endemic presence of this sand fly. Lutzomyia maranonensis is also present in endemic areas of Ecuador, another region where known vectors of Carrion’s disease are not present.3,11
Multiple areas of Peru, Ecuador, and Colombia present endemic or even sporadic periodic transmission of B. bacilliformis in the absence of known vectors. It has been hypothesized that in addition to L. maranonensis, other Lutzomyia species are probably involved in disease transmission. In Peru, L. pescei was found during outbreaks in Cusco, and L. serrana accounted for 94% of sand flies collected during an outbreak in a high jungle area of Huánuco.12 In addition, in an endemic region of northeastern Marañon, where known vectors were not reported, L. robusta and L. maranonensis accounted for 58% and 32% of all sand flies collected, respectively.11 In Ecuador, L. maranonensis has not only been previously described mainly in the Andean region (94.1%), bordering San Ignacio Province, one of the most relevant Peruvian areas of endemicity for Carrion’s disease, but also in Amazonian and Coastal regions (5.3% and 0.6%, respectively).19 Meanwhile, Lutzomyia columbiana and L. serrana could be potential vectors in endemic Colombian areas, where during the last decades, only sporadic cases have been reported.20,21 However, the presence of L. robusta, L. serrana, and L. maranonensis in Carrion’s disease–free areas could be considered as a risk factor which may lead to the introduction of the illness into these areas.
The conventional PCR assay used in this study was designed to detect Bartonella genus DNA but is also able to amplify phylogenetically related microorganisms such as Brucella spp.22 The conventional PCR detection of Mesorhizobium spp., nitrogen-fixing bacteria belonging to the order Rhizobiales, in L. maranonensis is not surprising. These microorganisms are closely related to Bartonella spp. and they have been found in the guts of different insects, including members of the Lutzomyia genus23–25 and blood-feeding invertebrates.26,27 Sand flies are known to be plant feeders, in addition to blood feeding during oviposition, and, for that reason, could obtain plant phyllosphere microbiota detectable with molecular and traditional bacteriological methods.26,27 In addition, the presence of other described/undescribed Bartonella spp., either pathogenic or not, cannot be ruled out. These data highlight the importance to accurately determine the presence of potential pathogens in vector-determining studies to avoid misidentifications.
The rate of B. bacilliformis sequencing confirmed in sand fly infection in our study was 0.52. Also, the individual positive rate in Querocotillo district was 0.98, similar to the value of 1.0 rate for L. peruensis in 1999.9 These results suggest that L. maranonensis could be an efficient vector similar to L. peruensis, but it is necessary to conduct further studies to validate this suggestion.
The lack of data regarding blood meal status of sand flies used in this study also needs to be considered, and it is an important limitation of the study. Because female sand flies can contain B. bacilliformis DNA after they have completed at least one blood meal, we cannot affirm whether the sand flies were infected or only fed from an infected source. Despite other limitations, such as a small sample size and a specific geographical study area, we were able to demonstrate the presence of B. bacilliformis DNA in L. maranonensis.
In summary, we show the presence of B. bacilliformis DNA in L. maranonensis, expanding the number of Lutzomyia species that could potentially transmit B. bacilliformis. Validation of this finding may explain the endemic presence of bartonellosis in certain areas of northern Peru and Ecuador, where known Lutzomyia vectors are absent.
Acknowledgments:
We are very grateful to Abraham Cáceres for his contribution in helping and supervising Lutzomyia species identification and Andrés G. Lescano for his critical review and statistical support of this manuscript. We are indebted to Pamela Sánchez V. for her support in the preparation of maps, and Carlos Palomares R. and Carmen Tinco V. for their support in the experimental phase of this study. We are also grateful to the collaborators of the Entomology Laboratory of National University of San Marcos, Lima—Peru. G. M. U. is an Emerge fellow, sponsored by the training grant D43 TW007393 awarded by the Fogarty International Center of the U.S. National Institutes of Health awarded to Emerge, the Emerging Diseases and Climate Change Research Unit of the School and Public Health Administration at Universidad Peruana Cayetano Heredia.
REFERENCES
- 1.Sanchez Clemente N, Ugarte-Gil CA, Solórzano N, Maguiña C, Pachas P, Blazes D, Bailey R, Mabey D, Moore D, 2012. Bartonella bacilliformis: a systematic review of the literature to guide the research agenda for elimination. PLoS Negl Trop Dis 6: e1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pachas PE, 2001. Enfermedad de Carrión (Bartonellosis) en el Peru. Lima, Peru: Ministerio de Salud, OGE, INS.
- 3.Minnick MF, Anderson BE, Lima A, Battisti JM, Lawyer PG, Birtles RJ, 2014. Oroya fever and verruga peruana: bartonelloses unique to South America. PLoS Negl Trop Dis 8: e2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pons MJ, Gomes C, del Valle-Mendoza J, Ruiz J, 2016. Carrion’s disease: more than a sand fly-vectored illness. PLoS Pathog 12: e1005863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomes C, Palma N, Pons MJ, Magallon-Tejada A, Sandoval I, Tinco-Valdez C, Gutarra C, del Valle-Mendoza J, Ruiz J, Matsuoka M, 2016. Succinyl-CoA synthetase: new antigen candidate of Bartonella bacilliformis. PLoS Negl Trop Dis 10: e0004989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomes C, Pons MJ, del Valle-Mendoza J, Ruiz J, 2016. Carrion’s disease: an eradicable illness? Infect Dis Poverty 5: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cáceres GA, Galati BAE, Pinto J, Paredes R, Reategui R, Perez J, Chevarría L, Yáñez H, Zorrilla V, 2000. Psychodidae (Diptera) del Perú I: phlebotominae en Huánuco, Pasco y Cusco, su relación con la enfermedad de Carrión y la leishmaniasis tegumentaria. Rev Peru Biol 7: 27–43. [Google Scholar]
- 8.Romero S, 2004. Detection of Bartonella bacilliformis by Real-Time PCR in Naturally Infected Sand Flies. Bethesda, MD: Faculty of the Department of Preventive Medicine and Biometrics Uniformed Services University of the Health Sciences.
- 9.Villaseca P, Padilla C, Ventura G, Samalvides F, Yañez H, Chevarría L, Ellis B, Rotz L, Leake J, Beati L, 1999. Importancia de la Lutzomyia peruensis en la transmisión de la enfermedad de Carrion en el Valle Sagrado de los Incas. Urubamba-Cusco, Peru. Rev Med Exp 15: 28–30. [Google Scholar]
- 10.Moo-Llanes DA, Arque-Chunga W, Carmona-Castro O, Yañez-Arenas C, Yañez-Trujillano HH, Cheverría-Pacheco L, Baak-Baak CM, Cáceres AG, 2017. Shifts in the ecological niche of Lutzomyia peruensis under climate change scenarios in Peru. Med Vet Entomol 31: 123–131. [DOI] [PubMed] [Google Scholar]
- 11.Caceres AG, Galati EA, Le Pont F, Velasquez C, 1997. Possible role of Lutzomyia maranonensis and Lutzomyia robusta (Diptera: Psychodidae) as vectors of human bartonellosis in three provinces of region nor Oriental del Marañon, Peru. Rev Inst Med Trop São Paulo 39: 51–52. [DOI] [PubMed] [Google Scholar]
- 12.Cáceres AG, Galati EAB, 2001. Lista de phlebotominae (Diptera: psychodidae) para el Perú y especies consideradas como vectores naturales e incriminadas en la transmisión de patógenos de la leishmaniosis tegumentaria y la enfermedad de Carrión (verruga peruana). Rev Med Exp 18: 100–106. [Google Scholar]
- 13.Ministerio de Salud , 2016. Boletin Epidemiológico del Perú. Volumen 25–Semana Epidemiológica N° 51. ISSN versión electrónica: 2415-0762. Available at: http://www.dge.gob.pe/portal/docs/vigilancia/boletines/2016/51.pdf. Accessed data May 1, 2018.
- 14.Municipalidad Provincial de Cutervo , 2015. Memoria Anual, 2015 Available at: http://www.municutervo.gob.pe/documentos/MEMORIA_ANUAL_2015.pdf. Accessed data May 1, 2018.
- 15.Young DG, Duran MA, 1994. Guide to the identification and geographic distribution of Lutzomyia sand flies in Mexico, the West Indies, central in South America (Diptera: Psychodidade). Mem Am Entomol Inst 54: 1–881. [Google Scholar]
- 16.del Valle Mendoza J, et al. 2014. Diagnosis of Carrion’s disease by direct blood PCR in thin blood smear negative samples. PLoS One 9: e92283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong-Mei L, et al. 2015. Detection of Bartonella bacilliformis by real-time PCR with TaqMan-MGB probe. Microbiol China 42: 427–435. [Google Scholar]
- 18.Paiva BR, Secundino NF, Nascimento JC, Pimenta PFP, Galati EAB, Andrade HF, Jr., Malafronte RS, 2006. Detection and identification of Leishmania species in field-captured phlebotomine sandflies based on mini-exon gene PCR. Acta Trop 99: 252–259. [DOI] [PubMed] [Google Scholar]
- 19.Gomez EA, Kato H, Hashiguchi Y, 2014. Man-biting sand fly species and natural infection with the Leishmania promastigote in leishmaniasis-endemic areas of Ecuador. Acta Trop 140: 41–49. [DOI] [PubMed] [Google Scholar]
- 20.Gomes C, Ruiz J, 2018. Carrion’s disease: the sound of silence. Clin Microbiol Rev 31: e00056–e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montoya-Lerma J, Cadena H, Segura I, Travi BL, 1999. Association of Lutzomyia columbiana (Diptera: Psychodidae) with a leishmaniasis focus in Colombia due to species of the Leishmania mexicana complex. Mem Inst Oswaldo Cruz 94: 277–283. [DOI] [PubMed] [Google Scholar]
- 22.Gomes C, Martinez-Puchol S, Pons MJ, Bazán J, Tinco C, del Valle J, Ruiz J, 2016. Evaluation of PCR approaches for detection of Bartonella bacilliformis in blood samples. PLoS Negl Trop Dis 10: e0004529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sant’Anna MRV, Darby AC, Brazil RP, Montoya-Lerma J, Dillon VM, Bates PA, Dillon RJ, 2012. Investigation of the bacterial communities associated with females of Lutzomyia sand fly species from South America. PLoS One 7: e42531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gouveia C, Asensi MD, Zahner V, Rangel EF, Oliveira SM, 2008. Study on the bacterial midgut microbiota associated to different Brazilian populations of Lutzomyia longipalpis (Lutz & Neiva) (Diptera: Psychodidae). Neotrop Entomol 37: 597–560. [DOI] [PubMed] [Google Scholar]
- 25.Kešnerová L, Moritz R, Engel P, 2016. Bartonella apis sp. nov., a honey bee gut symbiont of the class Alphaproteobacteria. Int J Syst Evol Microbiol 66: 414–421. [DOI] [PubMed] [Google Scholar]
- 26.Leulmi H, Bitam I, Berenger JM, Lepidi H, Rolain JM, Almeras L, Raoult D, Parola P, 2015. Competence of Cimex lectularius bed bugs for the transmission of Bartonella quintana, the agent of trench fever. PLoS Negl Trop Dis 9: e0003789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramírez-Puebla ST, Rosenblueth M, Chávez-Moreno CK, de Lyra MC, Tecante A, Martínez-Romero E, 2010. Molecular phylogeny of the genus Dactylopius (Hemiptera: Dactylopiidae) and identification of the symbiotic bacteria. Environ Entomol 39: 1178–1183. [DOI] [PubMed] [Google Scholar]