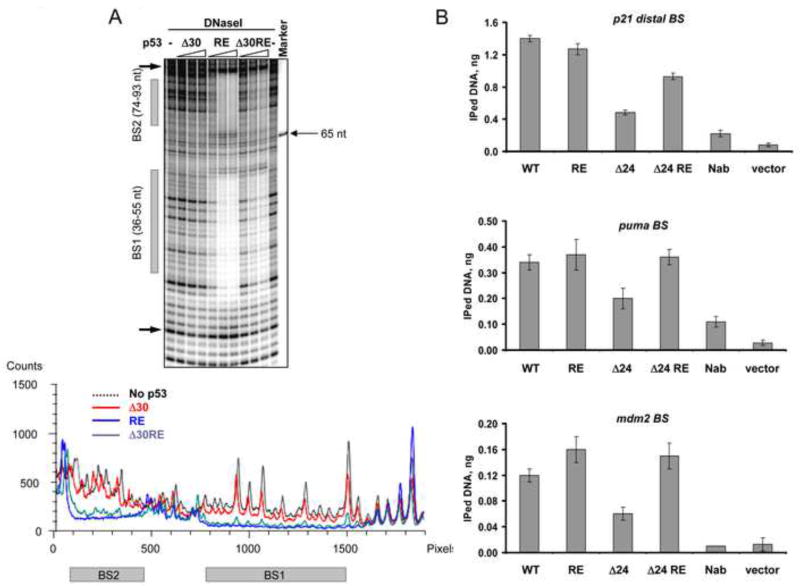
Figure 7. The E180R/R181E (RE) Double Mutation within the Central DNA Binding Domain Restores Interactions of CTD-Deleted p53 with Low Affinity Binding Sites in vitro and in vivo.
(A). DNaseI footprinting analysis was performed on 32P-labeled mdm2 DNA (non-template strand labeled) with either Δ30 p53, full length RE or Δ30 RE p53 proteins. On top is shown a portion of the PhosphorImager scan of the sequencing gel around the indicated p53 binding sites and the corresponding densitometry analysis specific to each p53 variant indicated by different colors at their maximal concentration is shown below. (B). Empty vector or constructs expressing full length WT, full length RE, Δ24 and Δ24 RE p53 proteins under control of the endogenous p53 promoter as indicated were transfected into H1299 cells. 24 h post- transfection the cells were subjected to the ChIP protocol (see Experimental Procedures). p53-DNA complexes were immunoprecipitated with MAbs 1801 and DO.1. p53 binding to its sites within the p21, puma and mdm2 promoters was evaluated by real time quantitative PCR and expressed as amount of immunoprecipitated DNA. See also Figure S7.