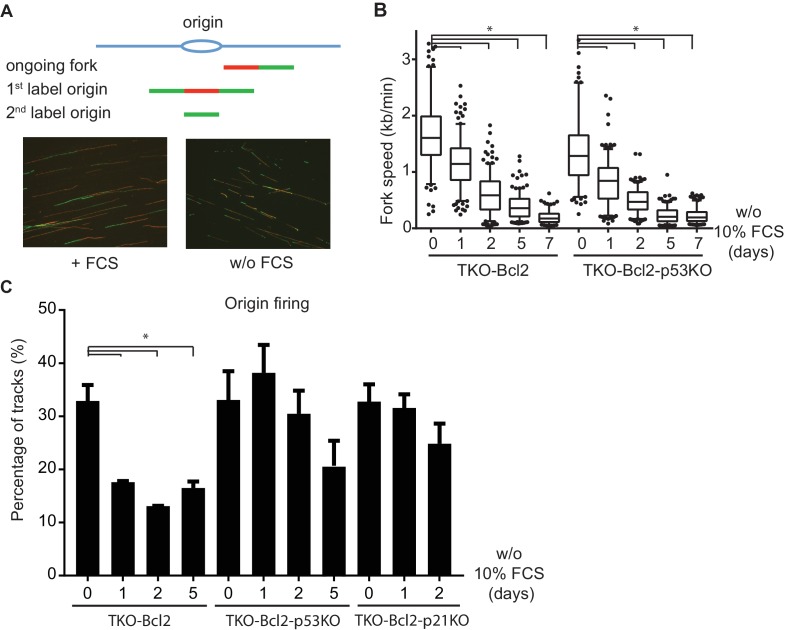
Figure 5. Loss of p53 restores the level of origin firing.
(A) Schematic representation of replication tracks generated after pulse labeling with CldU (red) and IdU (green). Ongoing forks were used to determine fork speeds (kb/min); 1st label and 2nd label origins are origins of replication initiated during the labelling period with CldU and IdU, respectively (upper panel). Representative images of DNA fibers of TKO-Bcl2 MEFs with and without 10% FCS (lower panel). (B) Replication fork speeds in TKO-Bcl2 and TKO-Bcl2-p53KO MEFs cultured in the presence or absence of 10% FCS for 1–7 days. Box plots represent interquartile ranges, horizontal bars denote the median, whiskers indicate 5–95 percentile and points are outliers. At least 350 track lengths of ongoing forks were measured (from three independent experiments) with ImageJ. Significant differences between median values are indicated with an asterisk (nonparametric Kruskal-Wallis test, p<0.05). (C) Quantification of origin firing in TKO-Bcl2, TKO-Bcl2-p53KO and TKO-Bcl2-p21KO MEFs cultured in the presence or absence of 10% FCS for 1–5 days. 1st label and 2nd label origins are shown as percentage of all labeled tracks (from three independent experiments). Significant differences between average values are indicated with an asterisk (p<0.05, Student’s t-test).