Abstract
Background:
Skeletal muscle mitochondrial content and function appear to be altered in obesity. Mitochondria in muscle are found in well-defined regions within cells, and they are arranged in a way that form distinct subpopulations of subsarcolemmal (SS) and intermyofibrillar (IMF) mitochondria. We sought to investigate differences in the proteomes of SS and IMF mitochondria between lean subjects and subjects with obesity.
Methods:
We performed comparative proteomic analyses on SS and IMF mitochondria isolated from muscle samples obtained from lean subjects and subjects with obesity. Mitochondria were isolated using differential centrifugation, and proteins were subjected to label-free quantitative tandem mass spectrometry analyses. Collected data were evaluated for abundance of mitochondrial proteins using spectral counting. The Reactome pathway database was used to determine metabolic pathways that are altered in obesity.
Results:
Among proteins, 73 and 41 proteins showed different (mostly lower) expression in subjects with obesity in the SS and IMF mitochondria, respectively (false discovery rate-adjusted P ≤ 0.05). We specifically found an increase in proteins forming the tricarboxylic acid cycle and electron transport chain (ETC) complex II, but a decrease in proteins forming protein complexes I and III of the ETC and adenosine triphosphate (ATP) synthase in subjects with obesity in the IMF, but not SS, mitochondria. Obesity was associated with differential effects on metabolic pathways linked to protein translation in the SS mitochondria and ATP formation in the IMF mitochondria.
Conclusions:
Obesity alters the expression of mitochondrial proteins regulating key metabolic processes in skeletal muscle, and these effects are distinct to mitochondrial subpopulations located in different regions of the muscle fibers.
Trial Registration:
ClinicalTrials.gov (NCT01824173)
Keywords: subsarcolemmal, intermyofibrillar, adiposity, mass spectrometry, proteome
1. Introduction
Mitochondria have a central role in energy metabolism in skeletal muscle, and several studies over the years have indicated impaired mitochondrial function in obesity and the associated insulin-resistant state. These studies have found reduced citrate synthase activity [1], reduced fatty acid oxidation [2], as well as lower capacity for mitochondrial electron transport [3] and ATP production [4] in the muscle of people with obesity/insulin resistance. Therefore, identifying differences in the abundance of certain proteins, or functionally related groups of proteins, in skeletal muscle mitochondria from lean subjects and subjects with obesity is imperative to get a deeper understanding of how obesity affects muscle mitochondrial function.
Although it is recognized that mitochondria form a reticulum within muscle cells, some of them are found beneath the sarcolemma, and they are called subsarcolemmal (SS) mitochondria; whereas the rest are found between the myofibrils, and they are called intermyofibrillar (IMF) mitochondria. The two mitochondrial subpopulations are biochemically distinct [5], and recent studies on rodents have provided evidence that individual types of mitochondrial proteins are differentially expressed between the SS and IMF mitochondria [6]. Current evidence suggests that the content of the SS mitochondria is regulated independently from that of the IMF mitochondria in individuals with obesity/insulin resistance [7]. Previous reports indicating differential expression of certain mitochondrial proteins in the skeletal muscle of humans with obesity [8, 9] have not distinguished between SS and IMF mitochondria. An obesity-associated reduction in mitochondrial enzyme activity is greater in the SS than in IMF mitochondria, and reduction in mitochondrial function in subjects with obesity is not entirely explained by a reduction in overall mitochondrial mass [10]. Such findings insinuate that the stoichiometry of mitochondrial proteins (i.e., abundance of individual proteins within a given mitochondrial mass) is altered in skeletal muscle of individuals with obesity depending on the subcellular localization of the mitochondria in the muscle fibers. Moreover, previous investigations employing animal models of metabolic impairments have found that Type 2 diabetes affects the expression of mitochondrial proteins in cardiac muscle differently in SS versus IMF mitochondria [11]. However, there are known differences in the mitochondrial proteome between skeletal and cardiac muscles [12]. To our knowledge, there is not yet evidence on how obesity may affect the proteome of SS mitochondria versus that of IMF mitochondria in human skeletal muscle.
Proteomic and associated bioinformatic analyses are powerful tools for understanding biological systems [13]. These investigative approaches can be used to explore population differences in the mitochondrial proteome and fundamental mitochondrial functions, ultimately leading to the discovery of potential therapeutic targets of energy metabolism within skeletal muscle. We aimed to compare differences in the abundance of individual proteins within skeletal muscle SS and IMF mitochondria between subjects with obesity and lean controls. To this end, we performed label-free quantitative proteomic analyses of SS and IMF mitochondria isolated from skeletal muscle of subjects with obesity and lean subjects. Furthermore, we performed bioinformatic analyses to obtain a deeper understanding of the metabolic implications of proteome differences in SS and IMF mitochondria in obesity. Our research provides novel insights into differential expression of mitochondrial proteins in muscle of humans with obesity and shows that these differences are distinct to the location of the mitochondria within the muscle fibers.
2. Materials and Methods
2.1. Participants
We studied 33 apparently healthy individuals, of which 17 had a BMI greater than 30 kg/m2 and 16 had a BMI less than 25 kg/m2. Statistical analysis based on differential expression of mitochondrial proteins in muscle homogenates from subjects with obesity and lean subjects [8, 9] indicated that 15 subjects per group would provide sufficient power (i.e., > 80%) for many mitochondrial proteins with >1 log2-fold difference between groups (FDR-adjusted P value of ≤ 0.05). Subjects were recruited through online and paper advertisements from the Greater Phoenix Metropolitan area in Arizona, USA, and assigned to the appropriate group based on their BMI. The experiments were conducted after obtaining approval from the Institutional Review Board at Mayo Clinic. The purpose, design, and risks associated with the overall experimental procedures were explained to each subject in the study before obtaining written consent, prior to the screening. All studies were performed in the CSIU at Mayo Clinic in Scottsdale, Arizona.
2.2. Screening and Muscle Biopsy
The screening procedures included medical history, routine physical examination, electrocardiogram, standard blood tests, and urinalysis. Insulin sensitivity was assessed by calculating the Matsuda ISI [14]. The HOMA-IR index was determined from the fasting plasma glucose and insulin concentrations. Subjects were excluded if they had diabetes; a history of liver, renal, or heart disease; or if they smoked, participated in a weight-loss regimen, took nutritional supplements, or used prescription or over-the-counter medications. Body composition was determined using BIA (310e, Biodynamics Corp., Shoreline, WA). Aerobic fitness was determined by measuring the maximal oxygen uptake during an incremental (20 W/min) cycle ergometer test to volitional exhaustion. A skeletal muscle biopsy (~ 100 mg) was collected from the vastus lateralis muscle on a separate day from the screening procedures, after an overnight fast, and after refraining from any form of exercise for 3 days before the biopsy procedure.
2.3. Isolation of Mitochondria and Mass Spectrometry
SS and IMF mitochondria were isolated from the muscle samples following procedures we have previously described [15] (Supplemental Figure 1). Protein content in the mitochondrial preparations was determined by the method of Lowry [16]. Skeletal muscle mitochondrial proteomic analyses were performed using an in-solution protein digest protocol for the generation of tryptic peptides and following established procedures [17, 18] we have also previously described [19].
All mass spectrometry analyses were performed by a member of the research team who was blinded during the analyses of samples. For the mass spectrometry analyses, 6 μL of 0.1% formic acid (v/v) was added to re-suspend the dried peptide samples before analyses of the samples by HPLC-ESI-MS-MS using a Thermo Scientific™ Orbitrap Elite™ Hybrid Ion Trap-Orbitrap Mass Spectrometer [19]. A “top 15” data-dependent MS/MS analysis was performed (acquisition of a full scan spectrum followed by collision-induced dissociation mass spectra of the 15 most abundant ions in the survey scan). Spectra were acquired using Xcalibur software (Thermo Fisher Scientific).
2.4. Database Searching
Database searches and criteria for protein identification were performed following procedures we have previously describing in detailed [19]. Briefly, tandem mass spectra were extracted using MSConvert script using the default parameters, and the fragment mass spectra were then searched against the human SwissProt_2017_01 database, using Mascot (Matrix Science, London, United Kingdom; version 2.4) and X! Tandem. Scaffold (version_4.6.1, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at a greater than 95.0% probability by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at a greater than 95.0% probability and contained at least two identified peptides. The ProteinProphet algorithm assigned all protein probabilities [20]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters.
2.5. Mitochondrial Protein Quantitation for Comparative Proteomics
Only proteins identified as mitochondrial by GO:0005739 were included in the analyses [21]. GO annotation was performed using the DAVID Bioinformatics Resources v6.7 (https://david.ncifcrf.gov/) [22] to manually curate proteins to functional groups (i.e., complexes I-V, TCA cycle).
Missing spectral count values across each of the identified mitochondrial proteins were replaced with the lowest spectral count value within each subject group of the lean subjects or the subjects with obesity [23]. Spectral counts for each protein were divided by the protein length to provide a SAF, and individual SAF values were further divided by the sum of all SAFs corresponding to the mitochondrial proteins within the sample to obtain a NSAF [8, 9]. A complication associated with the analysis of proteomic data is the occurrence of batch effects resulting from the analysis of large number of samples using high-throughput technologies [24]. Batch effects can result from differences in day-to-day laboratory conditions, reagents/equipment used, and technical personnel [25]. Therefore, and to optimize our comparative proteomic analyses, data were batch mean-centered to correct for variation between runs, and according to procedures previously described [24]. First, the mean NSAF value for a given protein within each run/batch was subtracted from each NSAF value for that protein in the same run/batch [26]. This resulted in negative values for some proteins, and therefore means for proteins within run/batch were then shifted to the protein batch median NSAF values calculated before the mean centering, and by adding the median NSAF value before mean centering to each batch mean-centered NSAF value [24]. Few proteins had still negative NSAF values, and to correct for that NSAF values were shifted to a minimum value of 0 spectral count by subtracting the minimum NSAF value observed for each protein across subjects [24]. These batch-corrected NSAF values were then used for statistical analyses. Only mitochondrial proteins detected in at least 50% of subjects (i.e., ≥ 17 of the 33 subjects), were used for comparisons between groups, a commonly employed threshold in similar proteomic studies [8, 9, 27, 28]. Supplemental Figures 2 and 3 detail the workflow related to the quantification of DEP between groups in SS and IMF mitochondrial fractions, respectively.
2.6. Bioinformatic Analyses of Differentially Expressed Mitochondrial Proteins Identified in SS and IMF Mitochondria
We used the Reactome pathway database (https://reactome.org/) [29, 30] to uncover metabolic pathways significantly enriched with DEP in obesity in SS and IMF mitochondria. Visualization of clustering of data using PCA and heatmaps was performed in the same data sets using ClustVis software (https://biit.cs.ut.ee/clustvis/) [31]. The STRING 10.5 database (https://string-db.org/) [32] was used to determine biological processes associated with the DEP in obesity in the SS and IMF mitochondria.
2.7. Statistical Analyses
Measurements between lean and obese groups were compared using Student unpaired t tests. The t tests were two-sided, and the level for significant difference was set at P ≤ 0.05. Discovery of DEP between groups was performed using the two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli, with a FDR of 0.1, and differences between groups with an FDR-adjusted P-value (i.e., q value) of ≤ 0.05 were considered significant [24, 33–35]. Data were mean-scaled to 1 for lean subjects when analyzing abundance of NSAF values of manually curated sets of proteins between groups. Data are presented as means ± SEM. Statistical analyses were performed using a commercially available software (GraphPad Prism version 7.00, GraphPad Software, La Jolla, CA).
3. Results
3.1. Proteomic Analysis of Isolated Mitochondrial Fractions
Anthropometric and metabolic characteristics of the individuals who met the study criteria and were included in the study are shown in Table 1. Per the study design, the two groups differed in BMI. As shown in Table 1, the subjects with obesity represented a typical population with obesity.
Table 1 –
Subject characteristics
| Lean | Obese | |
|---|---|---|
| n (F/M) | 16 (7/9) | 17(8/9) |
| Age (years) | 33.0 ± 2.5 | 31.5 ± 2.5 |
| Weight (kg) | 68.5 ± 2.6 | 98.5± 3.5*** |
| BMI (kg/m2) | 23.3 ± 0.6 | 34.2 ± 0.7*** |
| Body fat mass (%) | 22.5 ± 2.2 | 34.2 ± 1.6*** |
| FFM (kg) | 52.9 ± 2.4 | 64.8 ± 2.7** |
| VO2max (ml·min−1) | 2086 ± 164 | 2286 ± 134 |
| VO2max (ml·kgFFM−1·min−1) | 39.1 ± 2.0 | 35.3 ± 1.5 |
| Waist circumference (cm) | 80.1 ± 1.9 | 106.7 ± 3.2*** |
| Hip circumference (cm) | 98.1 ± 1.8 | 118.1 ± 2.6*** |
| Waist-to-hip ratio | 0.82 ± 0.02 | 0.91 ± 0.03** |
| Fasting plasma glucose (mg·dl−1) | 87.4 ± 1.6 | 91.4 ± 3.5 |
| Fasting plasma insulin (ulU·ml−1) | 5.0 ± 0.8 | 11.1 ± 1.2*** |
| HOMA-IR | 1.1 ±0.2 | 2.6 ± 0.3*** |
| Matsuda-ISI | 10.1 ± 1.5 | 4.2 ± 0.8** |
| HbA1c (mmol·mol−1) | 34.2 ± 0.5 | 36.2 ± 0.6* |
| HbAlc (%) | 5.2 ± 0.1 | 5.5 ± 0.1* |
| TSH (mIU·l−1) | 2.1 ± 0.4 | 2.4 ± 0.3 |
| Plasma triglycerides (mg·dl−1) | 86.6 ± 13.6 | 133.1 ± 20.1 |
| Total plasma cholesterol (mg·dl−1) | 170.9 ± 9.3 | 177.3 ± 6.5 |
| Plasma HDL-Cholesterol (mg·dl−1) | 61.3 ± 3.9 | 46.5 ± 2.9** |
| Plasma LDL-Cholesterol (mg·dl-1) | 92.3 ± 7.8 | 104.3 ± 6.3 |
Values are mean ± SEM. BMI, body mass index; FFM, fat-free mass; VO2max, maximal oxygen uptake; HOMA-IR, homeostatic model assessment of insulin-resistance; Matsuda-ISI, Matsuda insulin-sensitivity index; HbA1c, glycated hemoglobin; TSH, thyroid-stimulating hormone; HDL, high-density lipoprotein; LDL, low-density lipoprotein;
P ≤ 0.05,
P ≤ 0.01,
P ≤ 0.001 significantly different from lean subjects.
A total of 674 and 550 proteins were identified as mitochondrial in the isolated fractions designated as SS and IMF mitochondria, respectively. Among these mitochondrial proteins, 539 and 301 were detected in 17 of more subjects in the SS and IMF mitochondrial fractions, respectively (Supplemental Tables 1 and 2). Supplemental Figure 4 shows an area-proportional Venn diagram (BioInfoRx software (http://apps.bioinforx.com/;BioInfoRx, Inc., Madison, WI) of the identified mitochondrial proteins in the SS and IMF mitochondrial fractions, together with the proteins overlapping between the two mitochondrial subpopulations.
3.2. Abundance of Groups of Mitochondrial Proteins Associated with Key Biological Processes
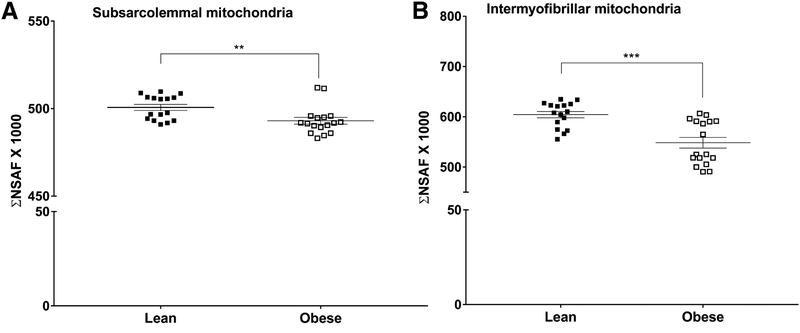
The ΣNSAF for the 539 identified mitochondrial proteins in the SS mitochondrial fraction was significantly reduced by 2% in subjects with obesity compared with the lean controls (P < 0.01; Figure 1A). The corresponding ΣNSAF for the 301 identified mitochondrial proteins in the IMF mitochondrial fraction was also significantly reduced by 9% in subjects with obesity compared with the lean controls (P < 0.001; Figure 1B).
Figure 1 –
Mitochondrial protein abundance in subsarcolemmal (A) and intermyofibrillar (B) mitochondrial fractions isolated from skeletal muscle of subjects with obesity and lean controls. Protein abundance is presented as the sum of normalized spectral abundance factors (ΣNSAF) of the mitochondrial proteins in each isolated mitochondrial fraction. Values are mean ± SEM. **P < 0.01, ***P < 0.001 versus lean.
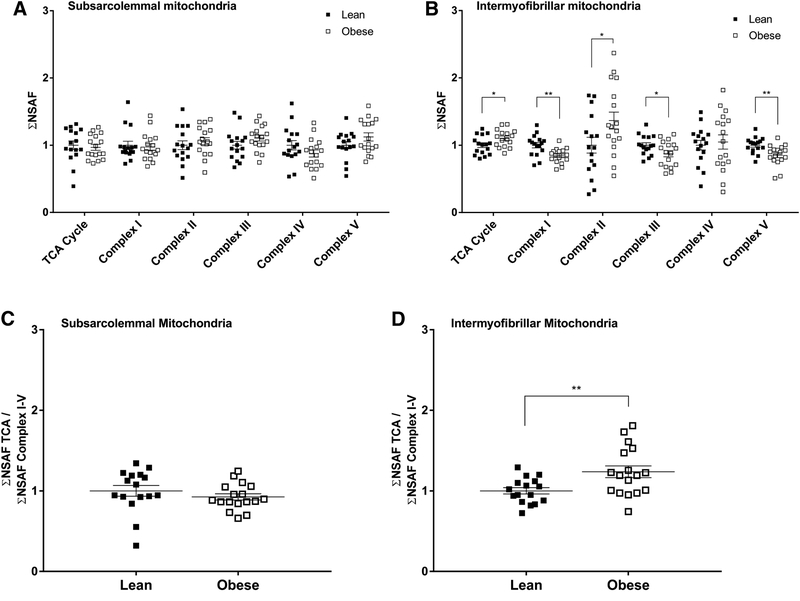
Manually curated groups of proteins assigned to the TCA cycle, the respiratory ETC (i.e., mitochondrial protein complexes I-IV), and ATP synthase had ΣNSAF that differed between subjects with obesity and lean controls in the IMF, but not SS, mitochondria (Figure 2A and 2B). The TCA cycle produces reduced coenzymes that are direct substrates for the ETC, ultimately contributing to the ATP production in the mitochondria. To evaluate the potential of generation relative to utilization of reducing equivalents as it relates to protein abundance, we calculated the ratio of ΣNSAF for the TCA cycle to that for the mitochondrial complexes I-V. As expected based on the data shown in Figure 2A, there was no difference between groups in the ratio of ΣNSAF for the TCA cycle to that for mitochondrial protein complexes I-V in the SS mitochondria (Figure 2C). However, this same ratio was significantly higher in the IMF mitochondria in subjects with obesity (P < 0.01; Figure 2D).
Figure 2 –
Abundance of manually curated groups of proteins assigned to the tricarboxylic acid (TCA) cycle, respiratory electron transport chain complexes, and ATP synthase (i.e., complex V) in subsarcolemmal (A) and intermyofibrillar (B) mitochondrial fractions isolated from skeletal muscle of subjects with obesity and lean controls. Abundance of proteins of the TCA cycle relative to that of complexes I-V in subsarcolemmal (C) and intermyofibrillar (D) mitochondrial fractions isolated from skeletal muscle of subjects with obesity and lean controls. Protein abundance is presented as the sum of normalized spectral abundance factors (ΣNSAF) of the respective groups of mitochondrial proteins in each isolated mitochondrial fraction. Values are mean ± SEM. All data have been adjusted to the lean group data for each variable. *P < 0.05, **P < 0.01 versus lean.
3.3. Metabolic Pathways Enriched with Differentially Expressed Mitochondrial Proteins in SS and IMF Mitochondria in Obesity
Among the 539 and 301 mitochondrial proteins identified in the SS and IMF subpopulations, respectively, 73 and 41 were significantly different between groups as determined by unpaired t test, and a FDR-adjusted cutoff P-value of 0.05. Among the 73 DEP in SS mitochondria, 28 and 45 were up- and downregulated, respectively, in the subjects with obesity (Table 2). Among the 41 DEP in IMF mitochondria, 13 and 28 were up- and downregulated, respectively, in the subjects with obesity (Table 3). Supplemental Tables 3 and 4 provide more details on the DEP in SS and IMF mitochondria, respectively.
Table 2 –
Differentially expressed mitochondrial proteins between lean subjects and subjects with obesity in SS mitochondria
| Gene Symbol |
log2 O/L Ratio |
q Value |
Gene Symbol |
log2 O/L Ratio |
q Value |
Gene Symbol |
log2 O/L Ratio |
q Value |
|---|---|---|---|---|---|---|---|---|
| ABHD11 | −1.84 | 0.0002 | SLC25A42 | −0.96 | 0.0044 | CASQ1 | 0.72 | 0.0458 |
| PDPR | −1.78 | 0.0001 | DHTKD1 | −0.93 | 0.0547 | IDH3A | 0.84 | 0.0036 |
| FDXR | −1.62 | 0.0001 | SLC16A1 | −0.92 | 0.0248 | TIMM21 | 0.85 | 0.0195 |
| ALDH2 | −1.53 | 0.0017 | TIMM44 | −0.92 | 0.0201 | ECHS1 | 0.85 | 0.0445 |
| NDUFB2 | −1.52 | 0.0195 | PDHA1 | −0.91 | 0.0044 | HK1 | 0.88 | 0.0302 |
| COMTD1 | −1.41 | 0.0065 | ATP5S | −0.89 | 0.0426 | IVD | 0.89 | 0.0160 |
| ALDH6A1 | −1.38 | 0.0001 | LDHD | −0.89 | 0.0316 | CAT | 0.92 | 0.0458 |
| OXCT1 | −1.34 | 0.0055 | MTX1 | −0.89 | 0.0354 | CYC1 | 1.01 | 0.0055 |
| ACAD9 | −1.34 | 0.0017 | FXN | −0.89 | 0.0103 | CISD3 | 1.04 | 0.0103 |
| MRPL4 | −1.31 | 0.0421 | MRPL45 | −0.84 | 0.0248 | APOOL | 1.06 | 0.0031 |
| MRPL2 | −1.27 | 0.0002 | ALDH4A1 | −0.83 | 0.0421 | AGMAT | 1.07 | 0.0082 |
| MTIF2 | −1.27 | 0.0081 | TIMM8A | −0.77 | 0.0458 | PARK7 | 1.08 | 0.0222 |
| BCKDK | −1.26 | 0.0547 | LRPPRC | −0.77 | 0.0082 | ME2 | 1.08 | 0.0025 |
| TBRG4 | −1.24 | 0.0003 | NDUFS1 | −0.75 | 0.0458 | PKM | 1.11 | 0.0012 |
| ACSS1 | −1.23 | 0.0080 | MRPS18B | −0.74 | 0.0080 | TMEM70 | 1.13 | 0.0017 |
| MIEF2 | −1.23 | 0.0153 | COQ9 | −0.72 | 0.0421 | SDHB | 1.13 | 0.0012 |
| MRPS30 | −1.22 | 0.0153 | NDUFB8 | −0.71 | 0.0195 | CNP | 1.17 | 0.0458 |
| ALDH7A1 | −1.14 | 0.0014 | ACOT9 | −0.71 | 0.0458 | MRPL28 | 1.18 | 0.0404 |
| MRPL18 | −1.10 | 0.0545 | HADH | −0.61 | 0.0142 | SCO2 | 1.19 | 0.0478 |
| DLST | −1.06 | 0.0017 | CYB5R1 | −0.56 | 0.0430 | GPX4 | 1.25 | 0.0458 |
| CCDC58 | −1.06 | 0.0377 | RDH13 | 0.52 | 0.0426 | COX17 | 1.55 | 0.0001 |
| BCKDHB | −1.04 | 0.0153 | IDH3G | 0.61 | 0.0547 | AK2 | 1.62 | 0.0017 |
| ECHDC3 | −1.03 | 0.0001 | ALDH5A1 | 0.61 | 0.0478 | CDS2 | 1.68 | 0.0044 |
| MPC1 | −1.02 | 0.0044 | CKMT2 | 0.64 | 0.0063 | |||
| HIGD1A | −0.98 | 0.0019 | NDUFB6 | 0.65 | 0.0190 |
O, obese; L, lean; q value, false discovery rate-adjusted P value.
Table 3 –
Differentially expressed mitochondrial proteins between lean subjects and subjects with obesity in IMF mitochondria
| Gene Symbol |
log2 O/L Ratio |
q Value |
Gene Symbol |
log2 O/L Ratio |
q Value |
Gene Symbol |
log2 O/L Ratio |
q Value |
|---|---|---|---|---|---|---|---|---|
| NDUFB2 | −2.01 | 0.0000 | ETFB | −1.05 | 0.0110 | CRAT | 0.55 | 0.0543 |
| PDK2 | −1.81 | 0.0000 | MAOA | −1.05 | 0.0275 | HIBCH | 0.56 | 0.0381 |
| CANX | −1.62 | 0.0038 | ALDH3A2 | −0.89 | 0.0390 | ATP5F1 | 0.62 | 0.0538 |
| HSPA5 | −1.59 | 0.0206 | PDP1 | −0.89 | 0.0217 | CS | 0.63 | 0.0543 |
| TIMM50 | −1.54 | 0.0201 | PRDX3 | −0.85 | 0.0243 | MT-ND5 | 0.76 | 0.0216 |
| ACSF2 | −1.32 | 0.0184 | CYC1 | −0.84 | 0.0538 | SLC25A20 | 0.95 | 0.0336 |
| MRPL21 | −1.29 | 0.0018 | LAP3 | −0.81 | 0.0234 | PITRM1 | 1.02 | 0.0234 |
| MRPL39 | −1.29 | 0.0058 | SUCLG2 | −0.79 | 0.0419 | ACSL1 | 1.04 | 0.0216 |
| NDUFB5 | −1.26 | 0.0025 | ALDH2 | −0.74 | 0.0111 | GATM | 1.06 | 0.0336 |
| ALDH6A1 | −1.22 | 0.0009 | ATP5J | −0.71 | 0.0217 | ACADM | 1.18 | 0.0217 |
| PHB | −1.16 | 0.0052 | ATP5F1B | −0.64 | 0.0025 | TRAP1 | 1.19 | 0.0217 |
| PGAM5 | −1.15 | 0.0354 | ACOT9 | −0.63 | 0.0201 | TMLHE | 1.21 | 0.0216 |
| NDUFA5 | −1.11 | 0.0052 | ALDH4A1 | −0.55 | 0.0216 | HDHD5 | 1.42 | 0.0333 |
| NDUFB6 | −1.06 | 0.0037 | ATP5FA1 | −0.49 | 0.0514 |
O, obese; L, lean; q value, false discovery rate-adjusted P value.
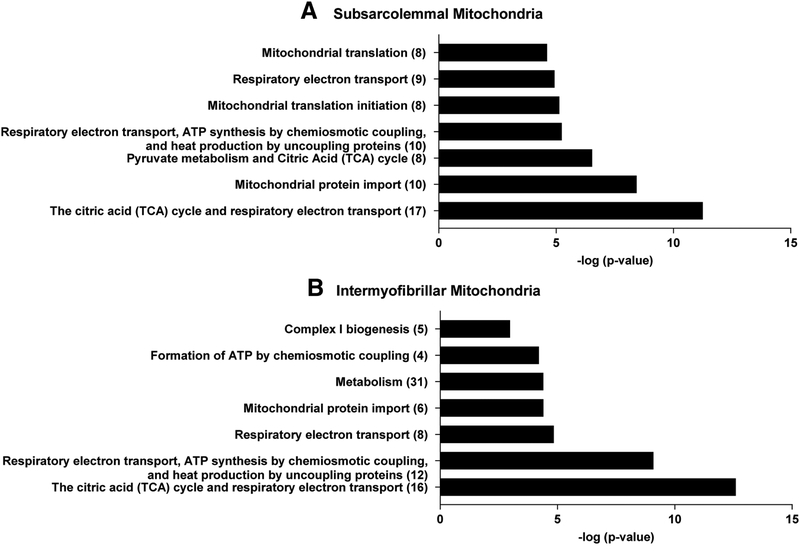
The seven most over-represented metabolic pathways enriched with DEP between groups in SS mitochondria are shown in Figure 3A, and a complete list of metabolic pathways enriched significantly with DEP in SS mitochondria based on Reactome analyses is included in Supplemental Table 5. Correspondingly, the seven most over-represented metabolic pathways enriched with DEP between groups in IMF mitochondria are shown in Figure 3B, and a complete list of metabolic pathways enriched significantly with DEP in IMF mitochondria is included in Supplemental Table 6. Characteristic mitochondrial proteins that were assigned to metabolic pathways showing differential protein enrichment in subjects with obesity versus lean controls in the SS and the IMF mitochondria are shown in Supplemental Figure 5.
Figure 3 –
Metabolic pathways enriched with differentially expressed proteins (DEP) between subjects with obesity and lean controls in subsarcolemmal (A) and intermyofibrillar (B) mitochondria based on Reactome pathway database analyses. The Y-axis represents the ranks of metabolic pathways based on P-values and the X-axis represents log-transformed false discovery rate P-value of enrichment test, with higher values representing more significant pathways. The number of DEP for each metabolic pathway is shown in parentheses. Tables showing the complete Reactome analyses for the subsarcolemmal and intermyofibrillar DEP between lean subjects and subjects with obesity are provided as Supplemental Tables 5 and 6, respectively.
Using the comprehensive protein-protein interaction network database STRING, we detected 56 protein-protein interaction networks in SS mitochondria associated with key biological processes as defined by GO and enriched with 5 or more of the 73 DEP (FDR adjusted P-value ≤ 0.05) (Supplemental Table 7). Corresponding analyses showed 24 protein-protein interaction networks enriched with the 41 DEP in the IMF mitochondria (Supplemental Table 8).
3.4. Organization of Differentially Expressed Proteins into Distinguishable Clusters in both SS and IMF Mitochondria in Obesity
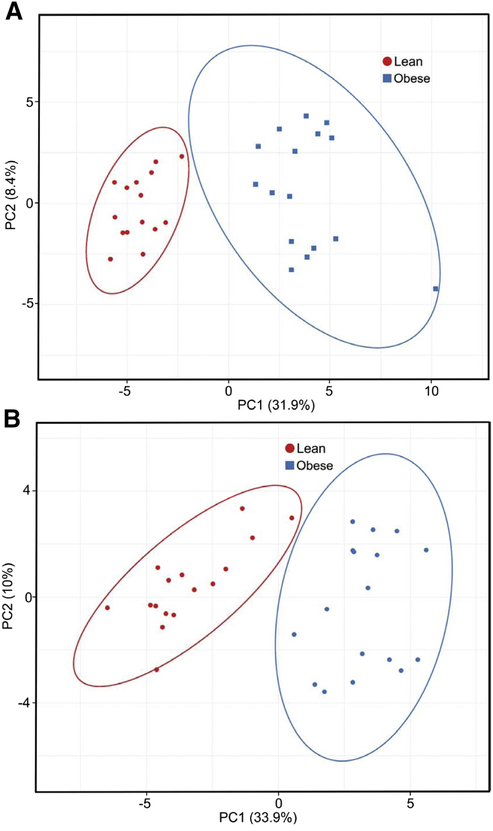
PCA successfully differentiated subjects with obesity from lean controls with respect to both SS (Figure 4A) and IMF (Figure 4B) mitochondria. Hierarchical clustering analysis using heatmaps of the same data showed that the DEP were organized into distinguishable clusters based on obesity status in both SS and IMF mitochondria (Supplemental Figures 6A and 6B).
Figure 4 –
Principal component (PC) analysis for differentially expressed subsarcolemmal (A) and intermyofibrillar (B) mitochondrial proteins in muscle of subjects with obesity and lean controls. X and Y axis show PC1 and PC2, respectively, that explain percent of the total variance. Prediction ellipses show clustering of the differentially expressed proteins between groups, and with probability of 0.95 that a new observation from the same group will fall inside the ellipse. N = 33 data points.
3.5. SS-to-IMF Expression of Individual Mitochondrial Proteins in Obesity
Among the mitochondrial proteins identified, 290 proteins were identified in both SS and IMF mitochondrial fractions from both subject groups. The overall mitochondrial protein abundance in SS mitochondria relative to that in IMF mitochondria (i.e., SS-to-IMF mitochondrial protein abundance ratio) describing the distribution of all 290 mitochondrial proteins within muscle was not different between the two groups (Supplemental Figure 7A). Similarly, the SS-to-IMF protein abundance ratios of manually curated groups of proteins assigned to key metabolic processes within the mitochondria were not different between the groups (Supplemental Figure 7B). On the other hand, Reactome pathway analysis using SS-to-IMF individual protein abundance ratios showed significant differential enrichment between groups in various metabolic pathways associated with the 290 mitochondrial proteins (Supplemental Table 9). Five proteins had SS-to-IMF protein abundance ratios that differed significantly between the subjects with obesity and the lean controls as determined by unpaired t-test and FDR-adjusted cutoff P-value of 0.05 (Supplemental Figure 8). Interestingly, the distributions of two proteins supporting immediate sources of energy in muscle as part of the phosphagen system (i.e., KCRS, mitochondrial creatine kinase; KAD2, adenylate kinase), were both higher in SS relative to IMF mitochondria in the muscle of subjects with obesity. Overexpression of mitochondrial creatine kinase is a compensatory response to impaired energy state associated with lower cellular phosphocreatine/ATP energy ratio [36].
4. Discussion
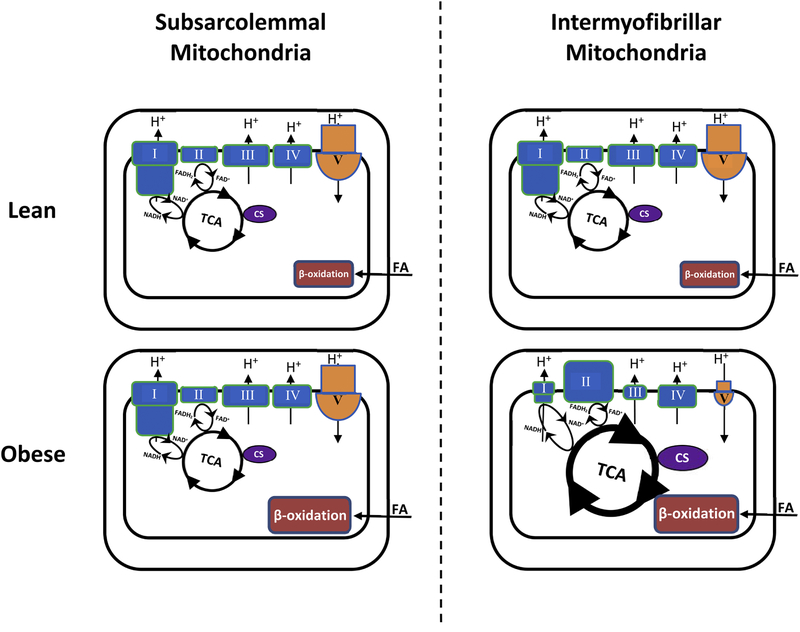
We show that the abundance of proteins of the ETC complexes I and III is lower in people with obesity along with that of the ATP synthase complex, but the abundance of proteins of the TCA cycle and ETC complex II is higher, and that these differences occur only in mitochondria located among myofibrils. On the other hand, mitochondria in the subsarcolemmal region from people with obesity show reduced abundance of proteins associated with protein translation. Figure 5 summarizes major proteome differences in energy metabolism we found predominantly in IMF mitochondria between subjects with obesity and lean controls. The overall findings demonstrate effects of obesity on the mitochondrial proteome that are distinct to the subcellular location of mitochondria in skeletal muscle.
Figure 5 –
Modeling of major proteome differences related to energy metabolism detected in skeletal muscle mitochondria between subjects with obesity and lean controls. The size of the symbols/letters in the graphs indicates higher or lower protein abundance in individuals with obesity (bottom) relative to that in lean controls (top) within a given muscle mitochondrial subpopulation. CS, citrate synthase; FA, fatty acids; TCA, citric acid cycle; I, complex I (NADH:ubiquinone oxidoreductaseI); II, complex II (succinate-coenzyme Q reductase); III, complex III (cytochrome c – oxidoreductase); IV, complex IV (cytochrome c oxidase); V, complex V (ATP synthase).
SS mitochondria from subjects with obesity showed reduction in DEP associated with mitochondrial translation (i.e., ribosomal proteins). Lower content of proteins involved in mitochondrial translation in the subsarcolemmal region of muscle cells will mediate reduced biogenesis of SS mitochondria previously reported in people with obesity/insulin resistance [37, 38]. On the other hand, the IMF mitochondria from subjects with obesity showed characteristic reduction in DEP associated with energy generation. They included proteins of the ATP synthase complex and ETC. From a functional standpoint, it appears that decreased content of proteins involved in mitochondrial respiration may not necessary impair mitochondrial function evaluated in vitro in animal models of insulin resistance [39]. Also, IMF mitochondria isolated from people with obesity/insulin resistance do not demonstrate impaired function in vitro [15]. Nevertheless, it still remains to be determined whether altered stoichiometry of mitochondrial proteins in IMF mitochondria, as seen in the present study, impairs ATP generation in vivo.
We found altered mitochondrial proteome in obesity with respect to metabolic pathways involved in fatty acid metabolism in both SS and IMF mitochondria. In SS mitochondria, we found an 80% increase in ECHS1 (i.e., enoyl-CoA hydratase) in subjects with obesity, and in association with differential regulation of the fatty acid β-oxidation pathway. These findings are comparable to that in rodents showing increased expression of proteins participating in the β-oxidation of fatty acids in SS mitochondria after high-fat feeding [40], or proteins involved in overall lipid metabolism in muscle of obese animal models [41]. In IMF mitochondria, we found an even greater increase (126%) in ACADM (i.e., acyl-CoA dehydrogenase) in subjects with obesity, an enzyme also involved in the β-oxidation of fatty acids. IMF mitochondria from subjects with obesity also showed increase in proteins involved in the activation of β-oxidation (i.e., ACSL1, long-chain-fatty-acid-CoA ligase 1, 105%) and transport of acylcarnitines across the mitochondrial membrane (i.e., CACT, carnitine/acylcarnitine translocase, 93%). Our study, therefore, suggests that at the mitochondrial proteome level the capacity to oxidize lipid is enhanced in people with obesity. In IMF, but not SS, mitochondria the increased capacity for β-oxidation of fatty acids was accompaniment by increased capacity at the level of the TCA cycle. It is noted that because of their proximity to the plasma membrane, SS mitochondria may have greater role in regulating processes related to insulin signaling in muscle [42]. Therefore, disproportionally greater capacity for β-oxidation of fatty acids relative to the capacity for utilization at the level of the TCA cycle specifically in SS mitochondria can lead to accumulation of fatty acid oxidation intermediates, such as acylcarnitines, which may interfere with the insulin sensitivity [43].
Lower abundance of mitochondrial complex I proteins in skeletal muscle is linked to higher oxidative stress [8]. Our combined findings about increased abundance in proteins comprising the TCA cycle and lower abundance in proteins found in the inner mitochondrial membrane in IMF mitochondria in people with obesity suggest a proteomic profile that has increased capacity to produce/dispose reducing equivalents of NADH and FADH2 in a less efficient ETC and generate oxidative stress. Given that IMF mitochondria comprise the majority of mitochondria in skeletal muscle (~80%) [44], increased oxidative stress generated by these mitochondria has important implications in terms of increasing the overall oxidative damage and may also impair insulin signaling [45]. We also found that the overall protein content of the ATP synthase is reduced in the IMF mitochondria. Lower content of proteins forming the ATP synthase complex is linked to lower ATP synthase activity [46]. Such response can increase the production of reactive oxygen species, as seen when the ATP synthase is experimentally inhibited in vitro by oligomycin [47]. Our overall findings at the mitochondrial proteome level suggest that the stoichiometry of proteins in mitochondria located among myofibrils is well-suited to induce oxidative stress in skeletal muscle of people with obesity.
We found that the protein content of the ATP synthase is reduced in subjects with obesity in the IMF mitochondria, which are responsible to support the energy needs of the muscle’s contractile activity [6, 42]. From a physiological perspective, decreased capacity for ATP synthesis in IMF mitochondria in individuals with obesity can decrease the capacity for physical activity [48], a phenomenon observed in people with obesity [49]. In turn, lower physical activity decreases the total daily energy expenditure and exasperates the weight gain seen in people with obesity [50]. Recently, structural alterations in muscle ATP synthase along with concomitant reduction in its activity were found to be key contributors to dyslipidaemia and insulin resistance in obesity [46]. Therefore, the protein content of the ATP synthase complex in IMF mitochondria emerges as a possible target for therapeutic interventions related to metabolic disorders in people with obesity.
Currently, there is considerable debate on whether BCAA are causally linked to insulin resistance [51]. Our Reactome pathway analyses showed significant enrichment with DEP of metabolic pathways related to the catabolism of BCAA in subjects with obesity compared with the lean controls in both SS and IMF mitochondria. ALDH6A1 (i.e., methylmalonate-semialdehyde dehydrogenase) involved in BCAA degradation decreased in subjects with obesity, in line with previous observations in whole muscle [8], in both SS and IMF mitochondria. Interestingly, BCKDK (i.e., branched-chain alpha-ketoacid dehydrogenase), which is the key regulatory enzyme in the inner mitochondrial membrane catalyzing the irreversible oxidative decarboxylation of branched-chain ketoacids [52], was reduced in subjects with obesity only in the SS mitochondria. Overall, our data suggest that impaired catabolism of BCAA in muscle may contribute to the elevated plasma BCAA concentrations in people with obesity, and that the subsarcolemmal region in muscle may constitute a site of considerable impairment in BCAA catabolism in these individuals.
4.1. Study Strengths and Weaknesses
We characterized skeletal muscle mitochondria in a subcellular-specific manner, which, when compared to previous investigations in muscle homogenates [8, 9], provides more detailed insight into the disturbances of the muscle mitochondrial proteome in obesity. Because obesity is a heterogeneous metabolic condition associated with a continuum of abnormalities [53], we used specific subject inclusion/exclusion criteria along with rigorous screening that allowed us to study homogeneous groups of subjects. We characterized these subjects extensively in terms of their physical and metabolic characteristics (Table 1), which enhances the reproducibility of our data. Moreover, we focused our studies in younger individuals to avoid well-characterized effects of aging on the muscle mitochondrial proteome [54].
A limitation of our study is that our subject population included only white participants, which constrains generalizability of our findings to other ethnic populations. A potential weakness of the experimental procedures used in the present study is the use of the proteolytic enzyme Nagarse, which is necessary to remove the myofibrillar proteins and liberate IMF mitochondria. Based on our previous work, treatment of muscle mitochondria with Nagarse also removes some mitochondrial proteins [19]. Therefore, some proteins associated with IMF mitochondria may not have been detected, and as a result, their expression could not be contrasted between groups. Also, because of the use of Nagarse in the IMF mitochondria, we could only compare data between mitochondrial fractions isolated under identical experimental procedures (i.e., we could not contrast data between SS and IMF mitochondria).
4.2. Study Conclusions and Translational Potential of the Findings
In conclusion, we found that the mitochondrial proteome in skeletal muscle of humans with obesity is altered in a subpopulation-specific manner. Specifically, proteins forming the TCA cycle were increased and those forming protein complexes of the ETC and the ATP synthase were decreased in subjects with obesity, but only in mitochondria embedded among the myofibrils. Obesity was associated with differential effects on metabolic pathways linked specifically to protein translation in the SS mitochondria and ATP formation in the IMF mitochondria. Because IMF mitochondria comprises the majority of mitochondria in skeletal muscle, altered proteome in IMF mitochondria can have considerable effects on skeletal muscle metabolism and, to that extent, on whole-body metabolism.
We identified proteins related to altered mitochondrial metabolism in obesity that are specific to the location of the mitochondria in skeletal muscle. Proteins found in the inner mitochondrial membrane of IMF mitochondria were largely reduced in people with obesity. In this regard, physical exercise, which has dominant role in increasing the content of IMF mitochondria [55], may serve as direct stimulus to normalize the content of proteins specifically in IMF mitochondria. Additionally, findings from this investigation may ultimately advance approaches related to the diagnosis of abnormalities in energy metabolism, improve the monitoring of mitochondrial disturbances, and serve as the testing ground for the critical application of lifestyle/drug regimens targeting the expression of mitochondrial proteins in skeletal muscle of people with obesity.
Supplementary Material
Highlights.
Stoichiometry of proteins is affected differently in SS vs IMF mitochondria by obesity
Proteins of complexes I and III are lower in obesity in muscle IMF mitochondria
Expression of TCA cycle proteins is higher in obesity in muscle IMF mitochondria
Expression of proteins of the ATPase is lower in obesity in muscle IMF mitochondria
Proteins regulating protein translation are reduced in obesity in SS mitochondria
Acknowledgments
We thank Dr. Wayne Willis for providing guidance and technical expertise related to the isolation of skeletal muscle mitochondria. We gratefully acknowledge the assistance of the subject recruitment and nursing staff with the conduct of the studies in the Clinical Studies Infusion Unit at Mayo Clinic in Scottsdale, Arizona. We also thank the subjects for their participation and commitment to the study procedures.
Funding
This work was supported by National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases grant R01DK094062 (CSK).
Abbreviations
- ATP
adenosine triphosphate
- BCAA
branched-chain amino acid
- BIA
bioelectrical impedance analysis
- BMI
body mass index
- CSIU
Clinical Studies Infusion Unit
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- DEP
differentially expressed proteins
- ETC
electron transport chain
- FADH2
reduced flavin adenine dinucleotide (reduced form)
- FDR
false discovery rate
- GO
gene ontology
- HOMA-IR
homeostatic model assessment of insulin-resistance
- HPLC-ESI-MS-MS
high-performance liquid chromatography-electrospray tandem mass spectrometry
- IMF
intermyofibrillar
- ISI
insulin sensitivity index
- NADH
nicotinamide adenine dinucleotide (reduced form)
- NSAF
normalized spectral abundance factor
- PCA
principal component analyses
- SAF
spectral abundance factor
- SS
subsarcolemmal
- STRING
Search Tool for the Retrieval of Interacting Genes/Proteins
- TCA
tricarboxylic acid
- ΣNSAF
sum of normalized spectral abundance factors
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
No conflicts of interest, financial or otherwise, are declared by the authors.
References
- [1].Machado MV, Ferreira DM, Castro RE, et al. Liver and muscle in morbid obesity: the interplay of fatty liver and insulin resistance. PLoS One 2012;7:e31738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Berggren JR, Boyle KE, Chapman WH, Houmard JA. Skeletal muscle lipid oxidation and obesity: influence of weight loss and exercise. Am J Physiol Endocrinol Metab 2008;294:E726–732. [DOI] [PubMed] [Google Scholar]
- [3].Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 2002;51:2944–2950. [DOI] [PubMed] [Google Scholar]
- [4].Abdul-Ghani MA, Jani R, Chavez A, Molina-Carrion M, Tripathy D, Defronzo RA. Mitochondrial reactive oxygen species generation in obese non-diabetic and type 2 diabetic participants. Diabetologia 2009;52:574–582. [DOI] [PubMed] [Google Scholar]
- [5].Palmer JW, Tandler B, Hoppel CL. Biochemical differences between subsarcolemmal and interfibrillar mitochondria from rat cardiac muscle: effects of procedural manipulations. Arch Biochem Biophys 1985;236:691–702. [DOI] [PubMed] [Google Scholar]
- [6].Ferreira R, Vitorino R, Alves RM, et al. Subsarcolemmal and intermyofibrillar mitochondria proteome differences disclose functional specializations in skeletal muscle. Proteomics 2010;10:3142–3154. [DOI] [PubMed] [Google Scholar]
- [7].Chomentowski P, Coen PM, Radikova Z, Goodpaster BH, Toledo FG. Skeletal muscle mitochondria in insulin resistance: differences in intermyofibrillar versus subsarcolemmal subpopulations and relationship to metabolic flexibility. J Clin Endocrinol Metab 2011;96:494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lefort N, Glancy B, Bowen B, et al. Increased reactive oxygen species production and lower abundance of complex I subunits and carnitine palmitoyltransferase 1B protein despite normal mitochondrial respiration in insulin-resistant human skeletal muscle. Diabetes 2010;59:2444–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hwang H, Bowen BP, Lefort N, et al. Proteomics analysis of human skeletal muscle reveals novel abnormalities in obesity and type 2 diabetes. Diabetes 2010;59:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ritov VB, Menshikova EV, He J, Ferrell RE, Goodpaster BH, Kelley DE. Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes 2005;54:8–14. [DOI] [PubMed] [Google Scholar]
- [11].Dabkowski ER, Baseler WA, Williamson CL, et al. Mitochondrial dysfunction in the type 2 diabetic heart is associated with alterations in spatially distinct mitochondrial proteomes. Am J Physiol Heart Circ Physiol 2010;299:H529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Forner F, Foster LJ, Campanaro S, Valle G, Mann M. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol Cell Proteomics 2006;5:608–619. [DOI] [PubMed] [Google Scholar]
- [13].Perakakis N, Yazdani A, Karniadakis GE, Mantzoros C. Omics, big data and machine learning as tools to propel understanding of biological mechanisms and to discover novel diagnostics and therapeutics. Metabolism 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care 1999;22:1462–1470. [DOI] [PubMed] [Google Scholar]
- [15].Kras KA, Hoffman N, Roust LR, Patel SH, Carroll CC, Katsanos CS. Plasma Amino Acids Stimulate Uncoupled Respiration of Muscle Subsarcolemmal Mitochondria in Lean but Not Obese Humans. J Clin Endocrinol Metab 2017;102:4515–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem 1951;193:265–275. [PubMed] [Google Scholar]
- [17].Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 2014;11:319–324. [DOI] [PubMed] [Google Scholar]
- [18].Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature protocols 2007;2:1896–1906. [DOI] [PubMed] [Google Scholar]
- [19].Kras KA, Willis WT, Barker N, Czyzyk T, Langlais PR, Katsanos CS. Subsarcolemmal mitochondria isolated with the proteolytic enzyme nagarse exhibit greater protein specific activities and functional coupling. Biochemistry and biophysics reports 2016;6:101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 2003;75:4646–4658. [DOI] [PubMed] [Google Scholar]
- [21].The Gene Ontology C Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 2017;45:D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- [23].Lazar C, Gatto L, Ferro M, Bruley C, Burger T. Accounting for the Multiple Natures of Missing Values in Label-Free Quantitative Proteomics Data Sets to Compare Imputation Strategies. Journal of proteome research 2016;15:1116–1125. [DOI] [PubMed] [Google Scholar]
- [24].Gregori J, Villarreal L, Mendez O, Sanchez A, Baselga J, Villanueva J. Batch effects correction improves the sensitivity of significance tests in spectral counting-based comparative discovery proteomics. J Proteomics 2012;75:3938–3951. [DOI] [PubMed] [Google Scholar]
- [25].Leek JT, Scharpf RB, Bravo HC, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nature reviews Genetics 2010;11:733–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lazar C, Meganck S, Taminau J, et al. Batch effect removal methods for microarray gene expression data integration: a survey. Briefings in bioinformatics 2013;14:469–490. [DOI] [PubMed] [Google Scholar]
- [27].Caruso M, Ma D, Msallaty Z, et al. Increased interaction with insulin receptor substrate 1, a novel abnormality in insulin resistance and type 2 diabetes. Diabetes 2014;63:1933–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xie X, Yi Z, Sinha S, et al. Proteomics analyses of subcutaneous adipocytes reveal novel abnormalities in human insulin resistance. Obesity (Silver Spring) 2016;24:1506–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fabregat A, Sidiropoulos K, Garapati P, et al. The Reactome pathway Knowledgebase. Nucleic Acids Res 2016;44:D481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Milacic M, Haw R, Rothfels K, et al. Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers 2012;4:1180–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res 2015;43:W566–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lundby A, Lage K, Weinert BT, et al. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell reports 2012;2:419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Simes RJ. An improved Bonferroni procedure for multiple tests of significance. Biometrika 1986;73:751–754. [Google Scholar]
- [35].Benjamini Y, Krieger AM, Yekutieli D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika, Volume 93, Issue 3, 1 September 2006, Pages 491–507 2006;93:491–507. [Google Scholar]
- [36].Schlattner U, Tokarska-Schlattner M, Wallimann T. Mitochondrial creatine kinase in human health and disease. Biochim Biophys Acta 2006;1762:164–180. [DOI] [PubMed] [Google Scholar]
- [37].Guillet C, Delcourt I, Rance M, et al. Changes in basal and insulin and amino acid response of whole body and skeletal muscle proteins in obese men. J Clin Endocrinol Metab 2009;94:3044–3050. [DOI] [PubMed] [Google Scholar]
- [38].Tran L, Kras KA, Hoffman N, et al. Lower Fasted-State but Greater Increase in Muscle Protein Synthesis in Response to Elevated Plasma Amino Acids in Obesity. Obesity (Silver Spring) 2018;26:1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lai N, Kummitha C, Hoppel C. Defects in skeletal muscle subsarcolemmal mitochondria in a non-obese model of type 2 diabetes mellitus. PLoS One 2017;12:e0183978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Dasari S, Newsom SA, Ehrlicher SE, Stierwalt HD, Robinson MM. Remodeling of skeletal muscle mitochondrial proteome with high-fat diet involves greater changes to beta-oxidation than electron transfer proteins in mice. Am J Physiol Endocrinol Metab 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Schonke M, Bjornholm M, Chibalin AV, Zierath JR, Deshmukh AS. Proteomics Analysis of Skeletal Muscle from Leptin-Deficient ob/ob Mice Reveals Adaptive Remodeling of Metabolic Characteristics and Fiber Type Composition. Proteomics 2018;18:e1700375. [DOI] [PubMed] [Google Scholar]
- [42].Hood DA. Invited Review: contractile activity-induced mitochondrial biogenesis in skeletal muscle. J Appl Physiol (1985) 2001;90:1137–1157. [DOI] [PubMed] [Google Scholar]
- [43].Koves TR, Ussher JR, Noland RC, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab 2008;7:45–56. [DOI] [PubMed] [Google Scholar]
- [44].Hoppeler H, Howald H, Conley K, et al. Endurance training in humans: aerobic capacity and structure of skeletal muscle. J Appl Physiol (1985) 1985;59:320–327. [DOI] [PubMed] [Google Scholar]
- [45].Fazakerley DJ, Minard AY, Krycer JR, et al. Mitochondrial oxidative stress causes insulin resistance without disrupting oxidative phosphorylation. J Biol Chem 2018;293:7315–7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Formentini L, Ryan AJ, Galvez-Santisteban M, et al. Mitochondrial H(+)-ATP synthase in human skeletal muscle: contribution to dyslipidaemia and insulin resistance. Diabetologia 2017;60:2052–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Roy A, Ganguly A, BoseDasgupta S, et al. Mitochondria-dependent reactive oxygen species-mediated programmed cell death induced by 3,3’-diindolylmethane through inhibition of F0F1-ATP synthase in unicellular protozoan parasite Leishmania donovani. Mol Pharmacol 2008;74:1292–1307. [DOI] [PubMed] [Google Scholar]
- [48].Rogge MM. The role of impaired mitochondrial lipid oxidation in obesity. Biol Res Nurs 2009;10:356–373. [DOI] [PubMed] [Google Scholar]
- [49].Cooper AR, Page A, Fox KR, Misson J. Physical activity patterns in normal, overweight and obese individuals using minute-by-minute accelerometry. Eur J Clin Nutr 2000;54:887–894. [DOI] [PubMed] [Google Scholar]
- [50].Ravussin E, Lillioja S, Knowler WC, et al. Reduced rate of energy expenditure as a risk factor for body-weight gain. N Engl J Med 1988;318:467–472. [DOI] [PubMed] [Google Scholar]
- [51].Gannon NP, Schnuck JK, Vaughan RA. BCAA Metabolism and Insulin Sensitivity - Dysregulated by Metabolic Status? Mol Nutr Food Res 2018;62:e1700756. [DOI] [PubMed] [Google Scholar]
- [52].Adeva-Andany MM, Lopez-Maside L, Donapetry-Garcia C, Fernandez-Fernandez C, Sixto-Leal C. Enzymes involved in branched-chain amino acid metabolism in humans. Amino Acids 2017;49:1005–1028. [DOI] [PubMed] [Google Scholar]
- [53].Ahima RS, Lazar MA. Physiology. The health risk of obesity--better metrics imperative. Science 2013;341:856–858. [DOI] [PubMed] [Google Scholar]
- [54].Staunton L, O’Connell K, Ohlendieck K. Proteomic Profiling of Mitochondrial Enzymes during Skeletal Muscle Aging. Journal of aging research 2011;2011:908035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lundby C, Jacobs RA. Adaptations of skeletal muscle mitochondria to exercise training. Exp Physiol 2016;101:17–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.