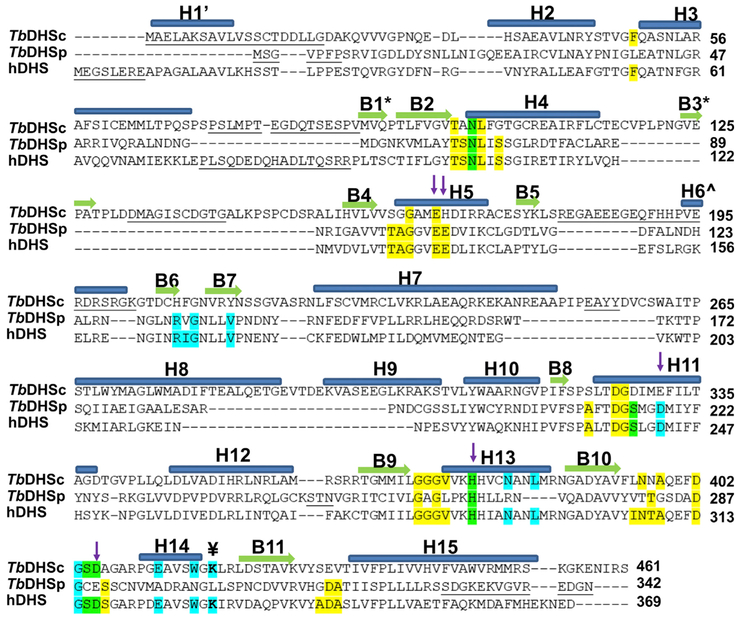
Figure 2. Structure-Based Sequence Alignment of hDHS (PDB: 1DHS), TbDHSc, and TbDHSp.
Residues highlighted show the NAD+ binding site (yellow), spermidine/GC7 binding site (cyan), and both ligands (green) from hDHS (PDB: 1RQD). The catalytic Lys is identified with the symbol ¥. Residues targeted for TbDHSc:DHSp mutagenesis are indicated with arrows. Helices are labeled with “H” and α sheets with “B.” B1* and B3* are found only in DHSc, H1’ is only in hDHS, and H6Λ^ is in both hDHS and DHSp. Structural alignment by PROMALS3D (Pei et al., 2008). Residues not resolved in the structure are underlined.