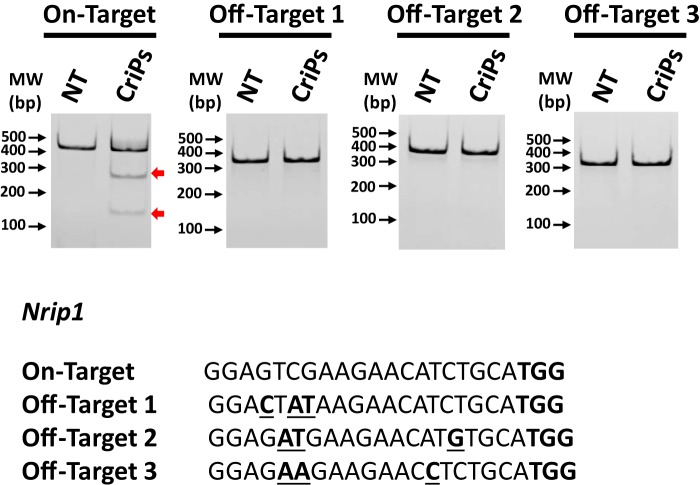
Figure 8.
Determination of off-target effects of CriPs targeting Nrip1 by a T7E1 assay. Primary pre-adipocytes were treated with CriPs with Nrip1 sgRNA 3. Pre-adipocytes were then differentiated into mature white adipocytes. Top off-target candidate sites were determined by the CHOPCHOP program. Off-target effects were determined by a T7E1 assay. Expected DNA bands cleaved by T7E1 were as follows: on-target: uncut, 420 bp; cut, 270 bp + 150 bp; off-target 1: uncut, 386 bp; cut: 283 bp + 103 bp; off-target 2, uncut: 387 bp; cut, 229 bp + 158 bp; off-target 3: uncut, 352 bp; cut: 182 bp + 170 bp. TGG in boldface type, the PAM site. Mismatch sites are underlined and in boldface type.