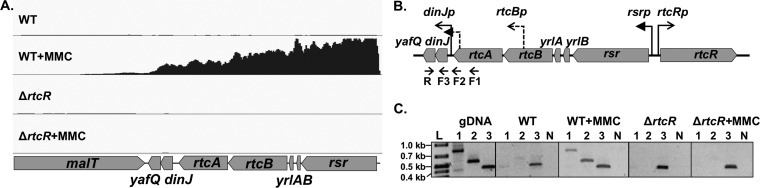
FIG 6.
Transcript analysis of RNA repair and dinJ-yafQ operons. RNA-seq data and endpoint RT-PCR analysis were used to assess the extent of the RtcR-dependent transcript initiated from rsrp. (A) RNA-seq data for the genome region carrying contiguous genes which showed RtcR-dependent upregulation are presented as reads mapped with Bowtie2 in the Integrated Genome Viewer (IGV). The y axis for each panel in the IGV reflects number of reads (range, 0 to 20,000 FPKM), and the x axis is the genome position (drawn to scale). One biological replicate of the two or three replicates used in the RNA-seq analysis is shown. (B) The RNA repair operon map is shown with the σ70-type promoter for the dinJ-yafQ operon, dinJp, and the potential promoters for RtcR-dependent expression of the dinJ-yafQ operon: the σ54-dependent promoter for the RNA repair operon (rsrp) and the Eσ54 binding site within rtcA. The reverse (R) and forward (F) primers for RT-PCR analysis of the dinJ-yafQ transcript(s) are indicated below the operon map. (C) For the RT-PCR analysis of the dinJ-yafQ transcript(s), RNA was isolated from MMC-treated and untreated cultures of WT and ΔrtcR strains and reverse transcribed with a gene-specific primer located in yafQ (R). cDNA was used in PCR with the R primer and forward primers F1, F2, F3, located in rtcA, overlapping the intergenic region downstream of rtcA, and in dinJ, respectively. PCR products from WT, WT+MMC, ΔrtcR, and ΔrtcR+MMC cDNA and from WT genomic DNA (gDNA) for the positive control were run on a 1.2% agarose gel. Lanes: L, 1-kb Plus ladder (Thermo Scientific); 1, F1/R primer pair; 2, F2/R primer pair; 3, F3/R primer pair; N, no-RT control with F3/R primer pair. Expected fragment sizes: F1/R, 812 bp; F2/R, 555 bp; F3/R, 424 bp. Results for one biological replicate are shown; two additional biological replicates gave identical results.