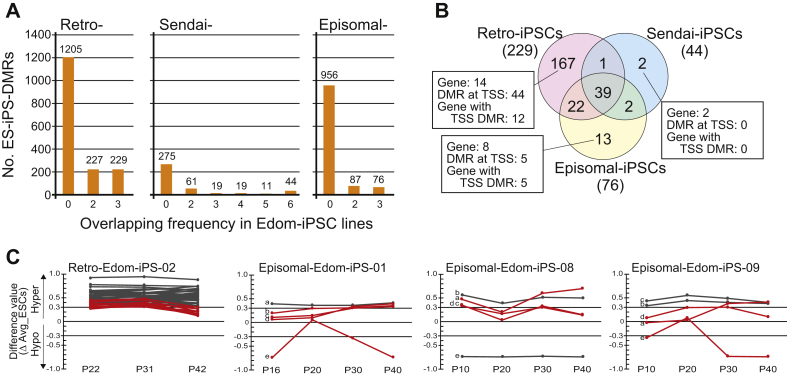
Fig. 3.
Aberrant methylation in human iPSCs. A, The number of overlapping ES-iPS-DMRs in 3 Retro-Edom-iPSC lines, 6 Sendai-iPSC lines and 3 Episomal-iPSC lines. B, Venn diagram showing method-specific DMRs among three kinds of vectors. C, Effect of continuous cultivation on 44 and 5 ES-iPS-DMRs at TSS in Retro-iPSC line and in Episomal-iPSC lines, respectively. The difference values were estimated by subtracting the scores of the averaged ESCs from that of each passage sample. Each line shows a difference value of each probe during culture. Red lines represent transiently ES-iPS-DMRs, which are not DMR at different passage and in different line. a, EPHA10 (probe ID: cg06163371), b, WIPF2 (cg04977733), c, FTH1 (cg25270670), d, ZNF629 (cg05549854), e, SLC19A1 (cg07658590).