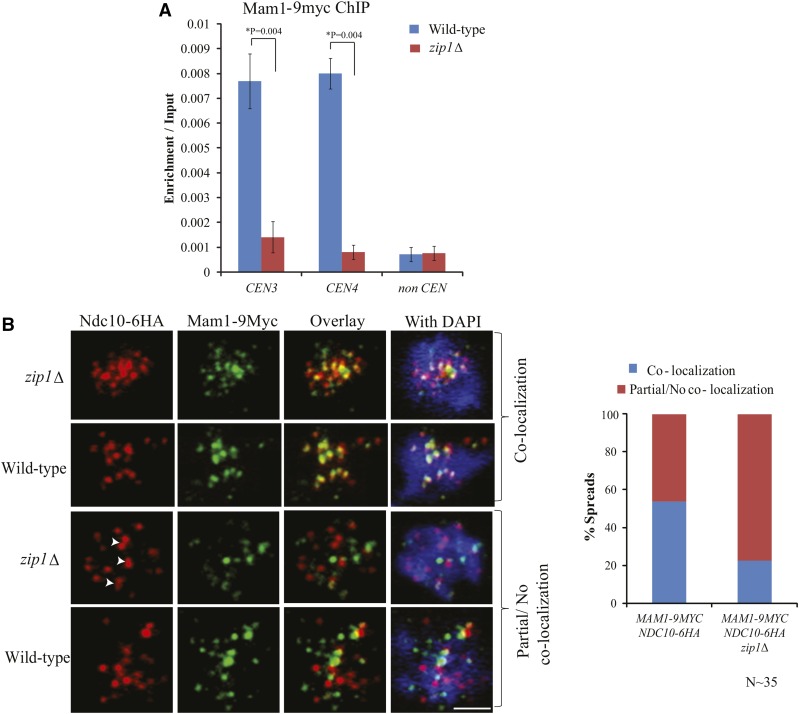
Figure 5.
Mam1 is mislocalized in the absence of Zip1. (A) ChIP assay for quantifying the association of Mam1-9Myc at the centromeres in the presence or absence of Zip1. Anti myc antibodies were used to pull down Mam1-9Myc. The experiment was repeated for three times and error bars represent the standard deviation from the mean. *The P value was determined by using ‘two tailed t-test assuming unequal variances’ option from the data analysis tool of Microsoft Excel. (B) Chromatin spread showing localization of Mam1-9Myc and Ndc10-6HA in prophase/metaphase I cells of wild-type (SGY1444) and zip1Δ (SGY1410). Arrowhead shows doublet of Ndc10 foci that signifies kinetochores remained together by flanking crossover in the absence of Zip1 (Gladstone et al. 2009). On the basis of the localization of Mam1-9Myc with respect to Ndc10-6HA, the cells were divided into two categories: co-localization, partial or no co-localization. Bar, ∼5 µm.