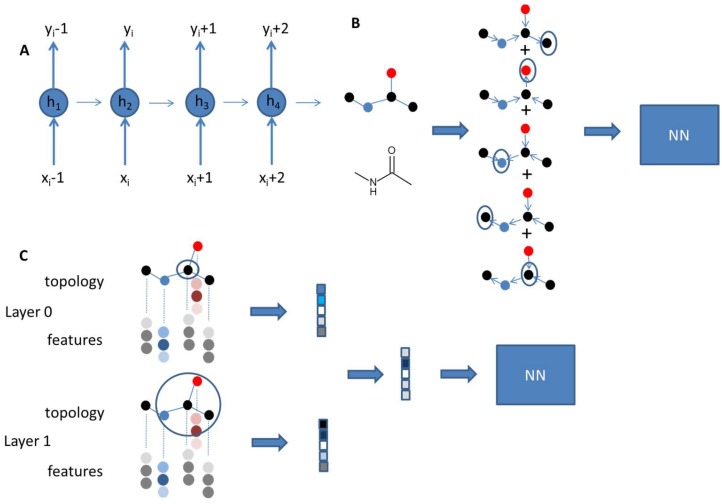
Figure 3.
(A) Recurrent neural networks (RNNs) use sequential data. The output for the next element depends on the previous element. Thus, RNNs have a memory. hi represent the hidden state at each neuron. They are updated based on the input x and the hidden state from the previous neuron. (B) In the UG-RNN approach, molecules are described as undirected graphs and fed into a RNN. Each vertex of a molecular graph is selected as a root node and becomes the endpoint of a directed graph. Output for all nodes is traversed along the graph until the root node is reached. All signals are summed to give the final output of the RNN, which enters into the NN for property training. (C) Graph convolutional models use the molecular graph. For each atom a feature vector is defined and used to pass on information for the neighboring atoms. In analogy to circular fingerprints different layers of neighboring atoms are passed through convolutional networks. Summation of the different atomic layers for all atoms results in the final vector entering the neural network for training.