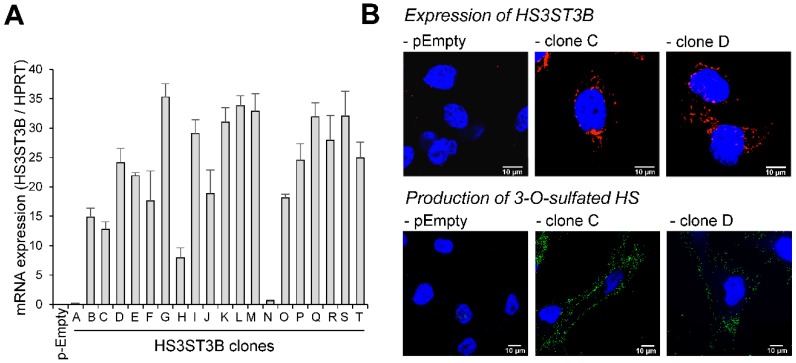
Figure 1.
Stable expression of HS3ST3B in MDA-MB-231 cells. Cells were transfected with the expression vector encoding HS3ST3B and then cultured in complete DMEM medium in the presence of 400 µg/mL G418. After 14 days of culture, individual colonies were isolated by limit dilution and amplified in medium supplemented with G418. In parallel, cells were transfected with the empty vector to obtain the control parental cells (pEmpty). (A) Following RNA extraction, the mRNA level of HS3ST3B was quantified by real-time RT-PCR in each clone. Relative abundance of the transcripts was normalized to endogenous HPRT mRNA. Data are means ± SD of triplicates. (B) HS3ST3B expression in the clones C and D was analyzed by confocal microscopy. To this end, cells were seeded on glass coverslips, permeabilized and then incubated in the presence of anti-HS3ST3B antibodies. After wash, they were immunostained with secondary antibodies conjugated to Alexa-568, in order to highlight the enzyme in red fluorescence. For the detection of 3-O-sulfated motifs, recombinant HSV-1 gD (10 μg/mL) was incubated with primary anti-gD antibody for 30 min at 4 °C, and the immune complex was added to cells for an additional 30 min-incubation. After washing, cells were fixed and incubated for 1 h with Alexa 488-conjugated secondary antibody (green fluorescence). In all the microscopy experiments, nuclei were stained in blue with DAPI, in order to visualize cell nuclei (N = 3 separate experiments; n = 30 cells). Scale bar = 10 µm.