Fig. 3.
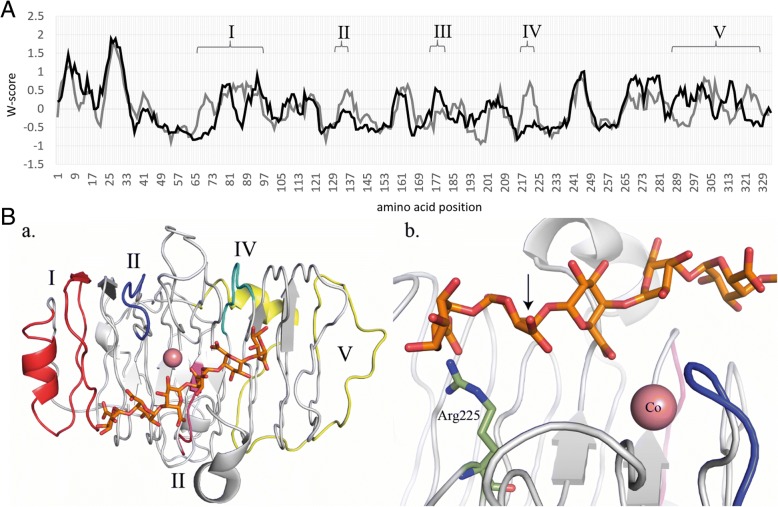
Reverse conservation analysis of PL1_7 orthologs revealed areas of putative functional divergence. a Amino acid conservation was estimated using Rate4Site, based on a MUSCLE alignment of fungal PL1_7 orthologs, and plotted as W mean scores in arbitrary units. The black and grey lines represent the closest orthologs in the PEL12/PEL13 and PEL2 clades (Additional file 2: Figure S1), respectively. b Cartoon representation of the PEL12 model. Defined regions are those that are mostly variable. I = pos. 67–98 (in red), II = pos. 131–137 (in blue), III = pos. 176–182 (in magenta), IV = pos. 217–223 (in cyan), V = pos. 287–327 (in yellow). The pentagalacturonate ligand is shown in orange sticks, and the two calcium ions positioned at two putative Ca2+ binding site, are shown in pink. In subfigure b. the catalytic base, Arg 225, is shown in green sticks. The hydrolytic cleavage of the polygalacturonic substrate will occur at the position denoted by the arrow. The pentagalacturonate and calcium ligands are extracted from a superposition of PDB:ID 3KRG