Abstract
Histologic examination of tissues is central to the diagnosis and management of neoplasms and many other diseases, and is a foundational technique for preclinical and basic research. However, commonly used bright-field microscopy requires prior preparation of micrometre-thick tissue sections mounted on glass slides, a process that can require hours or days, that contributes to cost, and that delays access to critical information. Here, we introduce a simple, non-destructive slide-free technique that within minutes provides high-resolution diagnostic histological images resembling those obtained from conventional haematoxylin-and-eosin-histology. The approach, which we named microscopy with ultraviolet surface excitation (MUSE), can also generate shape and colour-contrast information. MUSE relies on ~280-nm ultraviolet light to restrict the excitation of conventional fluorescent stains to tissue surfaces, and it has no significant effects on downstream molecular assays (including fluorescence in situ hybridization and RNA-seq). MUSE promises to improve the speed and efficiency of patient care in both state-of-the-art and low-resource settings, and to provide opportunities for rapid histology in research.
High-quality tissue microscopy is central to the diagnosis and management of neoplasms as well as other diseases. However, the bright-field (transmission) design of clinical microscopes requires optically thin (4–6 μm) slices of tissue mounted onto glass slides. Preparation of these slides is costly, generates toxic reagent waste, exhausts small samples, and perhaps most importantly, involves delays of hours to days that can adversely affect patient care as well as the efficiency of basic and preclinical research. Preparing frozen, as opposed to formalin-fixed, paraffin-embedded (FFPE) sections of fresh tissues, is a more rapid alternative but requires considerable expertise and the results are often unsatisfactory 1.
Optical alternatives that can acquire cellular-scale images directly from tissue surfaces without micro- or cryosectioning are under development. Approaches include structured illumination 2; conventional reflectance and fluorescence confocal microscopy 3; multi-photon imaging 4; spectrally encoded confocal microscopy 5; stimulated Raman microscopy 6 light-sheet microscopy 7,8; and optical coherence tomography 9, among others. While all appear promising, they have yet to be widely adopted by the cost-sensitive and technologically conservative field of pathology.
We present a simple and cost-effective, fluorescence-based, slide-free optical imaging system: MUSE (Microscopy with UV Surface Excitation). MUSE images can resemble those from standard histology slides and brightfield microscopes while also providing surface and colour contrast features not usually present in standard H&E preparations. The approach relies on two phenomena. The first: ultraviolet (UV) excitation light, particularly at wavelengths shorter than 300 nm, penetrates tissue just a few microns deep, i.e., the approximate thickness of a typical histology slide. This property of light-tissue interaction had been appreciated in previous work using a tunable laser intended to examine wavelength-dependent native tissue autofluorescence for in-vivo diagnostic purposes. It was noticed that excitation with light below 300 nm generated images with dramatically improved contrast and sharpness 10,11. The second: excitation light in this same sub-300-nm spectral range can elicit bright emission from tissue specimens stained with conventional fluorescent dyes. Despite being excited in the relatively deep UV, these stains emit photons in the visible range. The visible-band signals can then be captured using simple-to-operate and inexpensive conventional glass-based microscope optics and either grayscale or colour cameras. Fortunately, some of the dyes with this favorable excitation-emission behavior proved to label tissue components with specificities resembling those of haematoxylin and eosin.
Since the excitation light is localized to within a few microns of the surface, tissue sectioning is not required for achieving a high-contrast subcellular-scale image. Tissues, either fresh or fixed, can be stained and imaged with MUSE (at 3–10 frames per second) within just a few minutes, compared to delays of hours or days associated with current methods. Using colour-mapping approaches, the original fluorescence images can be converted to mimic conventional H&E-staining, or alternatively, novel optimized colour display schemes can be used 12. Finally, MUSE is non-destructive, meaning that small biopsy specimens can be imaged and then submitted for additional downstream studies as necessary. Preliminary results and examples of MUSE in dermatology were recently described 13,14.
Based on experience to date exploring the MUSE approach, we discuss key elements of the optical design and describe the associated straightforward staining methodology. Examples of slide-free imaging of a variety of tissue specimens are provided, and these are compared to conventional H&E histology. In some instances, we show that MUSE can also generate images that contain information unobtainable using standard thin sections and brightfield microscopy.
Results
UV surface excitation
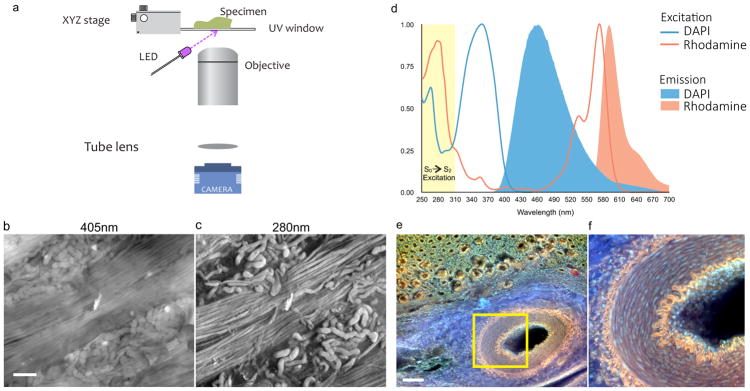
The optical system comprises one or more UV LEDs and UV-compatible sample stage, complemented by conventional microscope components (Fig. 1a). Oblique UV excitation light illuminates the specimen, bypassing the glass microscope lens, which, as it is opaque in the sub-300-nm spectral region, serves as an intrinsic excitation filter that blocks backscattered UV light from the optical path. The oblique excitation angle, as compared to full en-face illumination, can also generate shading across the face of a specimen that usefully highlights tissue surface topography (Fig. 1c). MUSE is distinct from other UV microscope systems (for example 15,16 which detect fluorescence emission from or absorbance by thinly sectioned samples, largely in the UV spectral range; unlike MUSE, such instruments require the use of special UV-transmitting or reflecting objective lenses.
Figure 1. Principles of MUSE: surface-weighted excitation and long Stokes-shift fluorescence emission.
a, Schematic diagram of MUSE optical design. An inverted microscope design is used, modified to include a UV-transparent stage and off-axis UV LED illumination. b,c, Unfixed ovine kidney, cut surface prepared via razor blade, stained with eosin and imaged with a grayscale camera, excited with c, 405-nm and d, 280-nm sources. Tubules and collecting ducts are easier to discern with 280-nm UV because of the latter’s surface-limited tissue penetration. d, 280-nm light excites multiple dyes that emit in the visible range due to the S0-S2 excitation (shown in yellow) followed by S1-S0 relaxation, excitation and emission spectra for DAPI and rhodamine are shown. e,f, Image captured from the cut surface of a thick, formalin-fixed porcine kidney specimen stained for 10 seconds with a mixed solution of rhodamine and Hoechst, and excited using a 280-nm UV LED. Multiple colours are visible: the muscular artery’s internal and external laminae (orange); collagen (blue); nuclei (lilac); tubules (orange and green). Endothelial cell nuclei extending down inside the artery are visible. Scale bars = 100 μm.
The advantage provided by sub-300-nm UV excitation compared to visible-range excitation is demonstrated in Figs. 1b and 1c. The cut surface of an unfixed lamb kidney specimen was stained with eosin (a fluorescent as well as chromogenic dye) and excited using either a visible-range, 405-nm LED (launched via traditional dichroic mirror with accompanying in-line emission filter) or an off-axis 280-nm UV LED. Images were captured with a monochrome CCD camera (see Methods for details) with the objective focused on the surface of the specimen. In this example, the 405-nm excitation generated an image with low overall contrast due to out-of-focus signals originating from multiple tissue depths. In comparison, 280-nm excitation of the same field provided a sharp, high-contrast image with clearly demarcated kidney tubules arranged among bundles of collecting ducts because the UV excitation was restricted to regions very close to the surface.
To identify optimal excitation wavelengths for MUSE, a tunable laser was used to illuminate stained cardiac tissue; images were acquired as the excitation was stepped from 350 nm to 210 nm. The results suggested that wavelengths from about 290 to 240 nm generate similar surface-limited excitation (Fig S1). See also Fig S2 and Supplementary Note 1 for further discussion.
280-nm UV excitation and standard fluorescent dyes
A variety of exogenous dyes were sought that would be excitable at 280 nm, emit in the visible range, and stain diagnostically relevant tissue features (nuclei, cytoplasm, and extracellular components) 17. Such behavior (UV-excitation with visible emission) would appear to be inconsistent with the narrow Stokes’ shifts usually associated with many fluorescent labels, but many visible-range fluorophores are in fact excitable at a less familiar, sub-300-nm spectral region, as illustrated with DAPI and rhodamine in Fig. 1d (see Supplementary Note 2 for additional discussion). Consequently, multiple stains can be combined, all excited at 280 nm while emitting across the visible range; the resulting colour signals are then captured simultaneously using a colour camera—of course, more sophisticated sensors can be used as well. As example, a kidney specimen stained for 10 seconds with a rhodamine-Hoechst solution is shown in Figs 1e and 1f. In addition to nuclear and parenchymal staining, the elastic laminae of the artery are visible in orange, while collagen demonstrates an intrinsic blue autofluorescence. Moreover, the oblique illumination and the extended depth of the unsectioned specimen provides a glimpse down into the vessel, the nuclei of the endothelial lining cells vanishing out of the field of view.
MUSE workflow compared to conventional histology
The steps in standard slide preparation involve fixation, orientation and trimming, dehydration (via alcohol and xylene), paraffin-embedding, block-facing and microtomy, transfer of sections to glass slides, rehydration, staining with haematoxylin and eosin, a second dehydration step, cover slipping, labeling, transport to a pathologist, or possibly to a whole-slide scanner, and review. While much of this is automated, it still requires hours and crucial input from highly skilled technicians. In contrast, MUSE processing minimally involves tissue orientation and possible surface-preparation via manual cutting using a sharp blade, staining tissue for as little as 10 seconds, rinsing for 20 seconds and placement directly onto a UV-transparent stage. Small, soft specimens can benefit from use of a mechanical jig to help orient the cuts. If the whole specimen does not have to be scanned, the resulting images can be viewed in real-time. Both fresh and fixed tissue specimens are suitable, and the resulting MUSE images are comparable suggesting that the fixation step and timing does not appear to be a crucial variable, at least for the dyes in current use. This is convenient since in many operating rooms tissues are automatically placed in formalin. In addition, samples acquired by non-planar cutting tools, such as the rounded blades of endoscopic biopsy forceps, can also yield acceptable results.
Colour conversion to virtual haematoxylin-and eosin-appearance
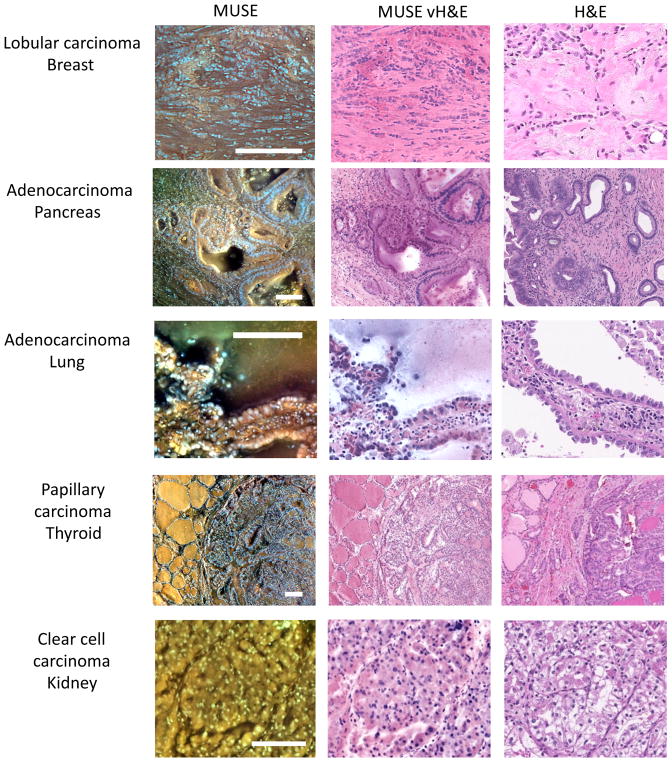
While MUSE captures images in fluorescence, histologists and pathologists are (currently) most comfortable looking at brightfield, H&E-stained specimens. In order to convert full-colour MUSE images (acquired in a single exposure using a colour camera) directly into virtual H&E (vH&E), a Python-language-based utility was developed that uses a variant of spectral unmixing and a Beer-Lambert blending model 18. A screenshot of this utility is shown in Fig. S3, see also Supplementary Note 3. Implemented on a GPU (graphics processing unit, code available on GitHub), such conversions can be performed automatically in near-real-time. As shown in Fig. 2, the conversion generates images that can closely approximate authentic H&E appearance and thus enhances image familiarity 19.
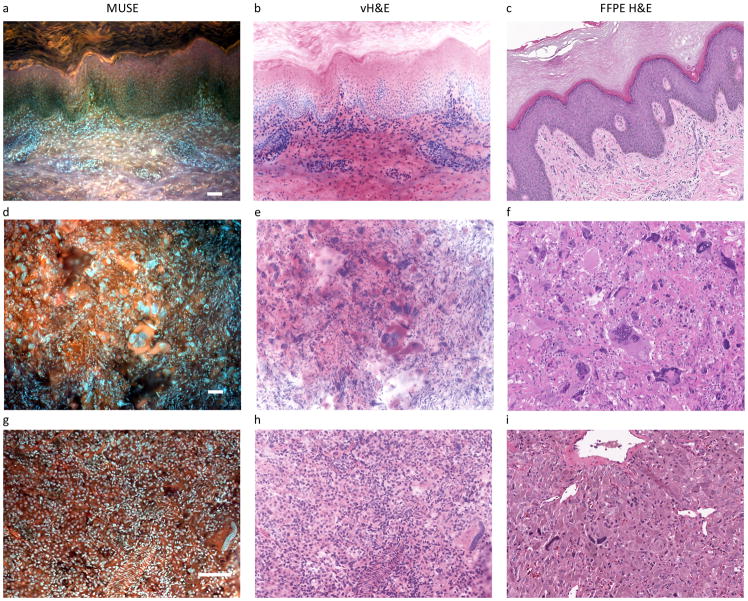
Figure 2. Fluorescence images, conversion to virtual H&E (vH&E) and comparison to paired FFPE conventional histology.
Left column consists of fluorescence MUSE images from the cut-surfaces of formalin-fixed tissues briefly stained with Hoechst, rhodamine, eosin and propidium iodide (PI), captured with a colour camera and white-balanced. Middle column: same images converted to vH&E as described in Methods and Supplementary Information. Right column, digital images captured with a whole-slide scanner from conventional H&E-stained slides of the same specimens after paraffin-embedding and sectioning. From top to bottom: lobular carcinoma, breast, with characteristic infiltration of single cells through stroma; adenocarcinoma, colon, with large glandular structures infiltrating the submucosa; adenocarcinoma, lung, with prominent lepidic spread of tumor cells along the alveolar lining; papillary carcinoma, thyroid, with clear distinction between normal and malignant regions; and clear cell carcinoma, kidney. Note absence of cytoplasmic clearing in MUSE images (compare with H&E). Scale bars = 100 μm.
Image quality and resolution, and suitability for replacing frozen sections
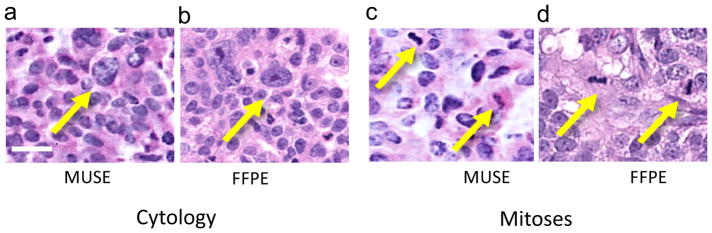
MUSE is capable of revealing important subcellular details, in particular, chromatin texture and mitotic figures. With a high-NA 10X lens, the current MUSE instrument generates imaging results that are close to that achievable with a conventional whole slide scanning operating at a nominal 20X setting, as shown in Figure 3.
Figure 3. High-resolution MUSE images for chromatin texture evaluation and mitosis detection.
Ovarian carcinoma (metastatic to the peritoneum), stained with Hoechst and rhodamine. a, Chromatin texture visible in MUSE image (colour mapped to vH&E); compare with b, FFPE H&E of a paired specimen. c, Mitoses are clearly visible and similar to those seen in d, FFPE paired specimen. Chromatin texture and mitoses are more readily appreciated after inverting the fluorescence image and conversion to brightfield mode. Scale bar = 20 μm
Frozen sections, used for intraoperative guidance, can be challenging to perform and often result in poor histology attributable to freezing artifacts 1. While MUSE promises to be suitable for replacing frozen sections, a formal assessment is still in progress; preliminary comparisons between MUSE, frozen sections and permanents are shown in Supplemental Figure S4. As with frozen sections, MUSE does not always faithfully reproduce some familiar and often useful artifacts attributable to FFPE processing 20. For example, the clear-cell carcinoma example in Fig. 3 does not show the eponymous cleared cytoplasm visible in the H&E image.
Extended field of view, or “whole-slide” imaging
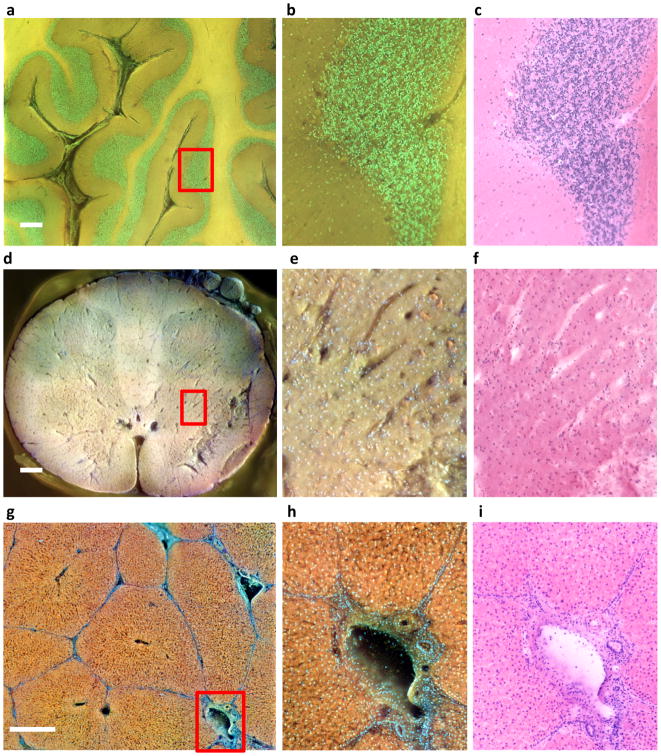
In regular histology practices, the size of the tissue to be imaged is limited by the capability of sectioning instrumentation; large-format microtomes, in use at a few sites, are expensive and technically challenging. However, experience suggests that large fields of view can provide insights not discernible with conventionally sized slides 21. A MUSE system equipped with a suitable stage can scan arbitrarily large specimens, for example, entire adult brain slices, with resolution uncoupled from sample dimensions. Examples of multi-field stitched images are shown in Fig. 4.
Figure 4. Large field of view imaging with MUSE.
Formalin-fixed tissue specimens (a–c, cerebellum; d–f, spinal cord; g–i, porcine liver) were imaged via MUSE. Flat surfaces were prepared by cutting with a hand-held histology blade and stained with rhodamine and Hoechst (cerebellum and liver), supplemented with eosin and PI (spinal cord). Multiple fields at 10X were captured using a scanning stage, and stitched together. The top row presents a region of cerebellum from a human neonate, consisting of 49 10X-images, flat-fielded and stitched together using the freely available Microsoft Image Composite Editor. b, Purkinje cells are visible at the interface between the molecular and granular layers. d–f, A 6 × 6 montage of a whole pediatric spinal cord cross-section is available at full resolution on line at http://www.gigapan.com/gigapans/199300.e, Neurons (orange) and capillaries are visible. g–i, An image montage (4 × 4 stitched 10X-images) of the cut-surface of a rhodamine-and-Hoechst-stained thick specimen of fixed porcine liver is shown in the bottom row. Normal liver architecture can be appreciated, with well-outlined lobules surrounding central veins and abutting portal triads consisting of portal veins, hepatic arteries, and bile ducts, are shown in h. (see http://www.gigapan.com/gigapans/185233.) Colour differences between these examples are due to intrinsic tissue properties and details of how colour-channel brightness and contrast were stretched. Scale bars = 700 μm for a and d, 300 μm for g.
Imaging speed is acceptable for collecting these large fields of view. Individual 10X images are currently captured at about 5 frames per second, which permits imaging a 15 × 15-mm region within about 2–3 minutes. Higher-power LEDs will shorten imaging time further. This scan time is comparable to that of conventional bright-field whole-slide scanners, with the proviso that the latter typically use effective lens magnifications of at least ~20X, providing somewhat higher spatial resolution than the 0.6 μm provided by the 10X, 0.45 NA-objective used in the present studies. While the current 10X objective can generate sub-nuclear resolution that may be adequate for most applications, MUSE systems deploying higher magnification objectives are technically feasible.
Preliminary validation studies
As with brightfield whole-slide imaging (WSI), recently cleared by the FDA after a process examining more than 2,000 specimens 22, MUSE will require detailed, multi-tissue, multi-pathology validation to support clinical use. A validation study was performed that compared diagnoses determined from MUSE images with those derived from viewing corresponding images from H&E-stained conventional slides of same specimens. The images were accompanied by tissue-of-origin information only. The study set consisted of 42 cases comprising both benign and malignant processes; the diagnoses made from MUSE and H&E images were essentially identical in 39 cases (a concordance rate of 93%). Of the 3 remaining cases, the paired diagnoses reflected alternate candidates on the differential diagnosis list, and the third discrepant case displayed diagnostic features in the MUSE images that were originally overlooked. Some representative example images are shown in Fig. 5, and case-by-case written diagnoses plus comments are provided in Supplemental table 1.. A separate study examining MUSE applicability to neuropathology cases with positive results is also presented in fig S5.
Figure 5. Sample images from the validation study with examples of concordant and discrepant diagnoses.
Columns as in Figure 2, except all MUSE-imaged specimens were stained only with rhodamine and Hoechst. Row 1 (Case #18--fixed): concordant. Benign skin with abundant keratin layer, easily diagnosed in both MUSE and H&E modes. Row 2 (Case #8—fresh): concordant. Retroperitoneal mass, easily identified as malignant neoplasm, and both reviewers favored rhabdomyosarcoma. Row 3: (Case 41—fixed): mildly discrepant. H&E reviewer favored adrenal cortical adenoma or carcinoma, MUSE reviewer favored pheochromocytoma, both part of a potentially difficult differential which may require IHC or other studies to resolve. Note large tubular nucleus in the MUSE image (lower right) and compare with similar long, dark nuclei in the corresponding H&E image. Scale bar = 100 μm.
Suitability for downstream molecular analysis
Increasingly, complex molecular diagnostic tests are being performed on ever smaller tissue samples, and a needle biopsy may have to be called upon to provide morphological diagnosis as well as molecular characterization. It would be ideal if MUSE could be used to non-destructively image such specimens (for diagnosis or sample adequacy determination) without interfering with downstream procedures, such as analysis of protein or DNA/RNA expression; targeted, exome or entire genome sequencing; metabolomics, and the like. It is also important that prior MUSE imaging not interfere with subsequent standard FFPE histology, IHC or FISH. Preliminary results presented in Supplemental Information suggest that prior MUSE imaging does indeed not interfere with these tasks (see Figs. S6–S8), although considerable further work is required to fully explore possible effects.
2.5-dimensional surface profiling and extended colour gamut
MUSE images can resemble conventional histology, but they also differ in several aspects. For example, 3-dimensional surface profile information can be appreciated, as shown in Fig. 6, a–f that is not easily perceived with the corresponding 5-μm-thin slices. Panels a and b demonstrate proteinaceous renal tubular casts; the MUSE image (acquired with a grayscale camera and emission filters and recoloured for clarity) clearly visualizes their cylindrical nature (unsurprising, but not easy to appreciate in the H&E). Panels c and d compare an H&E-stained preparation of a nerve sheath tumor with the MUSE image of the same specimen. The swirling nature of the tumor can be inferred from the H&E, but is more evident in the paired MUSE image. Finally, panels e and f examine the epithelium and fibrovascular core of a seromucinous ovarian carcinoma. An opening into a core can be seen in the upper right region of panel f. Additional examples of quasi-3D (thus termed 2.5D) views can be seen in Fig. 7. It is also possible to acquire actual quantitative depth information using Z-stack analysis (ms. in preparation). Interesting surface profiles imply excursions along the z-axis, which poses a challenge for high-magnification (high-NA) limited depth-of-field objectives. With Z-axis control it is possible to extend depth of field to ensure that entire specimens are in focus as shown in Fig. S9.
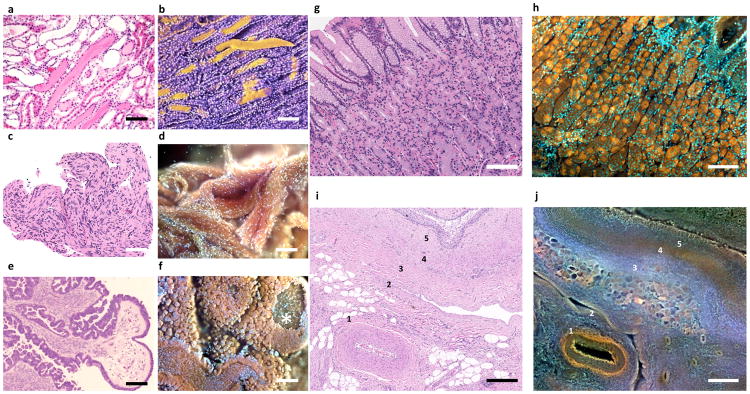
Figure 6. Additional shape and colour information available in MUSE vs. conventional H&E images.
a–f, Enhanced surface shape information. a, Standard FFPE-H&E histology of human kidney with proteinaceous casts filling some tubules. b, Same kidney imaged with MUSE, and pseudocoloured for clarity. Casts can be seen as cylindrical structures, including one that was dislodged from its original site. c, H&E Schwannoma. Swirling nature of this tumor can be appreciated. d, MUSE of same specimen, in which the 3D organization of the tumor can be appreciated. e, Seromucinous ovarian carcinoma, H&E. Carcinoma-lined tufts surround fibrovascular cores are visible. f, Same tumor imaged with MUSE, revealing cauliflower floret-like clumps of malignant epithelial cells outside fibrovascular cores. A window into a core is visible, top right.
g–j, Broader colour gamut reveals novel tissue contrast. g,h, Corresponding regions of normal human stomach fundus and i,j, porcine renal pelvis are shown, with MUSE specimens stained using rhodamine and Hoechst. g, Conventional FFPE H&E; h, MUSE, fluorescence image from a similar region. The chief (brown) and parietal cells (orange) are much easier to distinguish in the fluorescence MUSE image. i, Fixed porcine renal tissue, H&E; j, corresponding region, MUSE fluorescence mode. Stromal features, some identified by number, are easier to distinguish in the MUSE image vs. H&E. Scale bar = 100 μm.
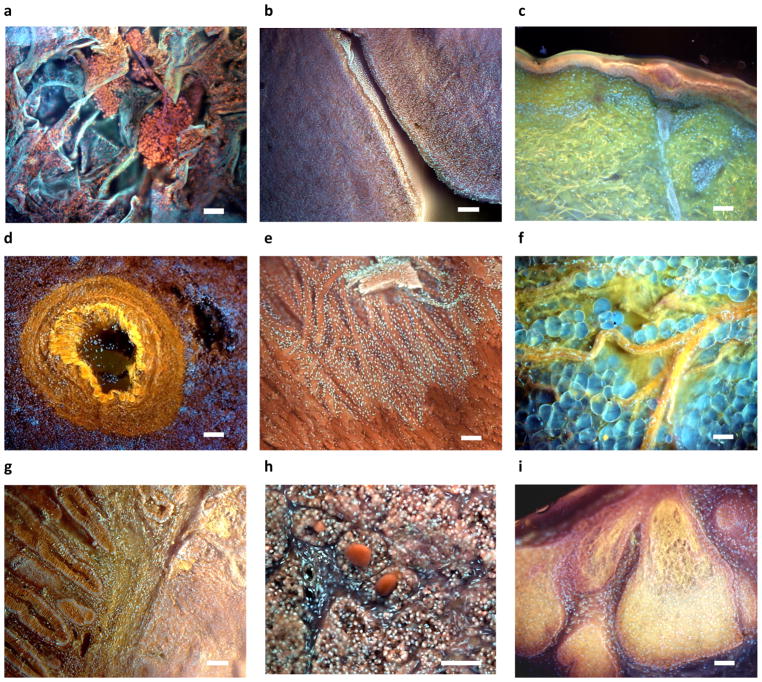
Figure 7. Examples of additional tissues imaged using MUSE.
a, Lung with clusters of histiocytes encased in a fine membrane (not visible on corresponding H&E images (eosin and PI); b, Ovary stroma and germinal epithelium c, Skin, with epidermis, dermis, skin appendages, collagen (green), elastin (yellow) and dermal-epidermal melanin visible (Hoechst, rhodamine, eosin, PI), d, Spleen, with prominent blood vessel (rhodamine and Hoechst); e, myocardium with thin layer of endocardium covering part of the specimen (rhodamine and Hoechst); f, fresh breast tissue, with nerve coursing over and through layer of intact adipocytes (rhodamine and Hoechst); g, Normal colon (left) next to adenocarcinoma (right)h, Pineal gland with pineal “sand” (rhodamine and Hoechst); i, sebaceous gland, cervix (rhodamine and Hoechst). Scale bar = 100 μm.
The staining repertoire available to MUSE is also broader than that seen with conventional H&E (Fig 6, g–j). For example, the stomach fundus contains two distinct cell types, chief and parietal cells. While these cell types are somewhat distinguishable in conventional histology, the two populations can be much more distinct following rhodamine and Hoechst staining (g vs. h). Stromal components that are hard to appreciate with conventional H&E can also be better distinguished with MUSE staining. In panels i (H&E) and j (MUSE) from a region of renal pelvis, features (labeled 1–5) are compared. The artery’s elastic laminae (1) and the vein (2), along with additional small vessels, are much easier to discern in the MUSE image. The three stromal layers (labeled 3–5) located beneath the urothelium in the upper right corner are tinctorially distinct with MUSE, but are only distinguishable in the H&E image through minor variations in texture. The stromal components reflected in these staining differences are not yet understood. The possible diagnostic utility of 2.5-dimensional views and extended colour ranges MUSE will require further investigation.
Additional examples of different tissue types imaged by MUSE
MUSE provides novel views of familiar tissues. Some examples of this, spanning a variety of specimens, are shown in Fig. 8. See figure legend for details, but some highlights include a remarkable cluster of histiocytes in lung (a), apparently surrounded by a thin purple sheath that also connects to other clusters (and not visible in paired FFPE specimens); multicolour view of skin with elastin, collagen and melanin clearly delineated (c); an unusual en-face view of endocardium with underlying myocardium (e); (f); nerve above and penetrating and between adipocytes from a fresh mastectomy specimen (f); breast ducts in stroma (g); skeletal muscle (h), and sebaceous gland (i), with tinctorial differences between the stem cells at the periphery and the maturing and apoptotic cells in the interior. Finally, it is possible to use MUSE in brightfield, transillumination mode to scan conventional whole slides, which could be a useful additional capability. An example is shown in Fig. S10.
Discussion
Although current microscope-based diagnostics are familiar and in use globally, they require sectioned and stained tissue slices mounted on glass slides, which may take hours to days to prepare and review. The overall process postpones diagnoses and contributes to overall health system inefficiencies and substantial, avoidable, patient anxiety. Hundreds of million slides are prepared just in the US each year, representing billions of dollars in technical costs alone. Conversely, rural areas and other low-resource settings may have no access to histology (and thus accurate diagnoses) whatsoever 23,24. In addition, as a microscopy technique alone, MUSE can contribute to fields as disparate as basic biology, microanatomy, toxicology, agriculture and education at all levels.
We demonstrate that 280-nm UV excitation, generated by now widely available LED sources, penetrates tissue to about the thickness of a typical tissue section, and generates images with excellent spatial resolution and contrast when tested on an array of normal and neoplastic tissues. It is possible to appreciate not only overall tissue architecture but also nuclear chromatin texture and mitotic figures. The samples are stained with inexpensive fluorescent dyes within seconds; resulting images can be colour-mapped in real-time to replicate standard H&E staining. The hardware itself is simple and robust, and the images are generated directly by a colour camera without requiring complex mathematical reconstruction. Preliminary validation studies described here indicate that MUSE indeed has the potential to generate images of diagnostic quality. Specimens ranging in size from needle biopsies to prostatectomies, and potentially to whole adult human brain slices, for example, can be accommodated simply by employing an XY stage with appropriate travel. Individual images can be acquired at 3–10 frames/second; enabling the capture of the equivalent of whole-slide scans in 2–3 minutes. Another important use case for MUSE is the rapid interrogation of large specimens to find critical areas to sample, either at higher magnification by MUSE and/or for selection of regions for FFPE, biobanking, or molecular testing. US CLIA regulations currently require the preservation of patient material even after diagnoses are rendered; following MUSE, tissues can be readily transferred into paraffin as necessary.
Both fresh and fixed tissues can be prepared and imaged using similar protocols. Fixed tissues are easy to orient and cut by hand, but their relative stiffness can interfere with having the cut face lie completely flat against the sapphire support. On the other hand, fresh tissue is more conformable, allowing small biopsies to be imaged directly without further surface preparation. However, it can be somewhat challenging to orient and section small soft specimens or ones with components of differing resistance to being cut by a blade. Jigs or temporary supports using fast-gelling agarose can help with cut-surface orientation.
In recent years, new treatment options that offer a personalized approach in treating cancer have been established. However, they require specific biomarker tissue testing to help predict response to new targeted therapy, sometimes years following primary diagnosis. Relatively non-invasive needle biopsies are useful for interrogating tumor recurrences especially in patients in advanced stages of disease who have failed several lines of conventional therapy. The non-destructive nature of MUSE can be particularly helpful in this setting, especially for small tissue biopsies that might otherwise be significantly consumed following frozen and/or conventional sectioning. Prior MUSE imaging does not appear to interfere with subsequent conventional histology, IHC, FISH or RNA-Seq studies. In fact, FFPE-based methods (dispensable with MUSE) can themselves have deleterious effects on nucleic acid integrity 25, antigen preservation and retrieval 26–29.
MUSE was developed with an eye towards examining manually cut tissue specimens, but there are tasks that call for examining closely spaced serial sections, for example, searching for small metastases in tumor-draining lymph nodes. Such fine spacing can be accomplished using vibrating blade microtomes able to slice fresh tissue at spacings of 50 μm or less 30. Another possible limitation: some useful artifacts associated with FFPE techniques, such as the chromatin clearing characteristic of some tumors, may not be visible in MUSE images, which in this regard resemble fine-needle aspirates or frozen sections 31.
Additional differences between MUSE and conventional histology exist, some advantageous, others perhaps may be distractions. (1) As UV penetration is slightly deeper than the thickness of conventional sections, cellularity estimates trend somewhat higher and nuclear crowding may interfere with some manual or automated evaluations. (2) A broader colour palette is available than that afforded by simple H&E. Even a simple two-dye cocktail (rhodamine and Hoechst), when combined with tissue autofluorescence (e.g., from collagen and tryptophan) or under the influence of other environmental effects 32, can generate multiple hues. On the one hand, such hues can serve as instant special stains in histology, highlighting important features such as elastic laminae that may be poorly delineated in H&E-staining. Converting these high-content colour images into H&E-like bright-field versions without a) confusing the viewer used to viewing a limited colour range, or b) collapsing the extra information into just 2 colour channels, remains an interesting challenge. (3) MUSE can generate 2.5-dimensional views of surface topography, revealing novel aspects of tissue organization and connectivity. While this may be an advantage in some settings, anything that diminishes the familiarity of the resulting images may be undesirable. That said, the prominence of topographical features in MUSE images can be regulated by how the surface of the specimen is prepared.
Methods
Optical design and components
As shown in Fig. S1 in the Supplementary Information section, the essential components of a functional system, based on an inverted microscope geometry, consist of a UV-transparent stage, one or more obliquely oriented 280-nm LEDs, and a conventional microscope optical train and imaging sensor. The specimen, which can be of any thickness, and stained as described below, is supported on 300-μm-thick, UV-transparent sapphire window (GT Advanced Technologies, Salem, MA, USA). The sample is oriented with a nominally flat surface, created in most cases using a knife or razor blade, towards the objective. The specimen can be lightly compressed against the stage to help flatten the tissue. The sapphire window is held in a custom mount attached to a motorized XYZ translational stage (433-series, Newport, Irvine, CA, USA) to allow for fully motorized translational scanning and focus.
The current version of the MUSE microscope prototype employs 280-nm UV-emitting LEDs for sample excitation (MTE280H32-UV, Marktech, Latham, NY, USA). These UV LEDs had a maximum output of 0.9 mW per LED. The light from one or more UV-LEDs was focused onto the surface of the sample using short-focal-length ball lenses in an oblique off-axis orientation, illuminating an approximately 1-mm2 area. Note: recently released LEDs in the same wavelength range (Nikkiso America, San Diego, CA, USA) can have maximum output of 30 mW or more.
The emitted fluorescent light was collected using a variety of long-working-distance objectives (e.g., 10X NA 0.28, Mitutoyo, Kawasaki, Japan, and 10X NA, 0.45 Nikon, Tokyo, Japan) and focused using an infinity-corrected tube lens (Thorlabs-ITL200, F = 200 mm, Thorlabs, Newton, New Jersey, USA) onto either a grayscale CCD camera (Retiga 2000 EXi, 1.5 megapixel, 6.4 μm/pixel, QImaging, Surrey, Canada), a Bayer-pattern colour CCD camera, either a Micro Publisher 3.3, 3.1-Mpixel, 3.45 μm/pixel (QImaging, Surrey, Canada), or a 9.2-Mpixel, 3.7 μm/pixel, colour CCD (Ximea, Münster, Germany). The grayscale camera was used to collect the dataset for exploring wavelength-dependent imaging depth, whereas the colour cameras were used to collect the other images shown (except for Fig. 6b). Using the 9.2-Mpixel colour camera and a 10X objective, the field of view comprised approximately 1 mm2.
In addition to the core components described above, the MUSE system was temporarily augmented with additional optics to allow for a comparison with conventional epifluorescence excitation (Fig. S1 in Supplementary Information); these included a 405-nm LED, a dichroic mirror (Thorlabs 500 LP, Thorlabs, Newton, New Jersey, USA) and a long-pass emission filter (Thorlabs FEL0500, Thorlabs, Newton, New Jersey, USA) To explore the wavelength-dependent depth of imaging in tissue, a laser system equipped with a tunable optical parametric oscillator (Vibrant 355I, Opotek, Carlsbad, California, USA) was used to generate excitation light in the 210-to-350-nm range. Mirrors were used to guide the laser light onto the sample with oblique geometry like that used by the LED excitation path, and the emitted fluorescence was collected as described above, except that a 500-nm long-pass filter was inserted to ensure that the entire range of excitation wavelengths used would be prevented from reaching the camera.
A convenient feature: the inverted optics design of this configuration allows the scanning of conventional whole stained slides in brightfield mode simply by using room light (or appropriately configured built-in white light sources) in transmission. See Supplementary Fig. S9 for an example image.
Assessment of depth of UV light penetration in biological tissue
To investigate the depth of penetration of the excitation light into tissue as a function of wavelength, a specimen of fresh porcine heart was stained for 16 hours (to allow deep penetration into the tissue) with Hoechst 33342 (Life Technologies, Carlsbad, CA, USA, 500 μg/ml in PBS) a dye that selectively labels nuclei, and the specimen was then imaged using tunable laser excitation, a blue long-pass filter, and a monochrome camera (shown in Supplementary Information Fig. S1). The excitation light was tuned from 350 to 210 nm in 10-nm steps. At each of the 15 wavelengths used, single-field images at 10 focal depths were collected by moving a 20X-objective lens using a micrometer stage in 4-μm steps.
The stack images were projected onto a single image using the maximum intensity Z-projection algorithm in ImageJ. Thereafter, a rolling-ball algorithm (ImageJ) was used to highlight the nuclei and suppress background signals resulted from excess binding of Hoechst to myocyte cytoplasm attributable to extended exposure to the stain. The nuclei were counted manually, assisted by an ImageJ cell-counter plugin.
Image acquisition and processing
Microscope control and image acquisition tasks were performed with custom software written in Microsoft Visual Basic .NET (VB.NET, Microsoft Corp, Redmond, WA, USA). Grayscale and colour images were captured with exposures that typically ranged from 0.1 to 0.5 seconds per frame, and were saved in TIFF or JPEG format. Image processing was performed using open-source ImageJ image processing and analysis software (http://imagej.nih.gov/ij/) along with open-source GIMP image processing software (https://www.gimp.org), and was in most cases confined to flat-fielding, adjusting brightness and contrast, colour balance, and sharpening with an unsharp mask tool (with the GIMP default settings, radius 5.0, amount 0.5, threshold 0.0). Using an XYZ translation stage (the Z axis is currently used for manual focus); multiple fields of view were collected and assembled by mosaicking single frames together. In many cases, the XYZ stage was sufficiently precise that the images were simply abutted after flat-fielding to create acceptable large montages. However, optimal montage results were obtained by allowing 10% image-to-image spatial overlap and using the Microsoft Image Composite Editor (http://research.microsoft.com/en-us/um/redmond/projects/ice/) to perform subpixel registration and image stitching.
When extended depth of field imaging was required, multiple Z-stacks were acquired at 10-micron spacing. See Supplementary Fig. S9 and Note 4 for more information.
Sample preparation and staining
In this study, both fresh and formalin-fixed tissues were used. Human specimens were sourced through two mechanisms. Predominantly, de-identified excess patient material was obtained directly from the frozen section or grossing room at UC Davis Medical Center. These tissue samples were determined to be exempt from oversight by the UC Davis Institutional Review Board (IRB ID 743439-1). Alternatively, formalin-fixed specimens from patients diagnosed with primary brain tumors were collected under IRB approval from the Department of Pathology and Laboratory Medicine at UC Davis Medical Center. Only excess tissues were retained and reviewed by a board-certified neuropathologist (co-author ML) for diagnostic purposes and for tissue quality control. Tissue samples were then coded and de-identified per protocol. Efforts were made to obtain tissue from both genders and from minorities. Animal tissues were obtained from the UC Davis Meat Lab from slaughtered animals, or from discarded experimental animals post-sacrifice; in the latter case, all procedures were performed under IACUC supervision.
Several dyes and dye-combinations were studied, including eosin, rhodamine, DAPI, Hoechst, acridine orange, propidium iodine, and proflavine. Eosin and rhodamine stain cytoplasm and the extracellular matrix, making the bulk of the tissue visible. Hoechst and DAPI fluoresce brightly when bound to DNA, allowing them to serve as excellent nuclear stains. A suitable combination proved to be rhodamine B (Sigma Aldrich, St. Louis, MO, USA 500 μg/ml in PBS) plus Hoechst 33342 (Life Technologies, Carlsbad, CA, USA, 500 μg/ml in PBS), which were combined in a single solution. Tissues were submerged in this combination for 10 seconds, and then briefly washed in water (fixed tissues), or in PBS (fresh tissues). The resulting stained tissue specimens generated bright enough signals for direct and interpretable visualization through microscope eyepieces, as the reddish rhodamine contrasts well with blue nuclear labels. Compared to captured digital images, live binocular viewing provides better appreciation of surface shape information. Camera-acquired images, however, benefit from additional digital enhancement, as described above.
For precise correlation between H&E and MUSE, we prepared conventional FFPE and H&E-stained slides, and then released the remaining tissue from the paraffin block by deparaffinization (essentially reversing the solvent steps used in embedding). The cut face of the recovered tissue specimen was then imaged via MUSE, creating essentially a serial-section rendition, as illustrated in Fig. S2. Alternatively, after primary MUSE imaging, samples were then processed for standard histology; the resulting H&E-stained slides were scanned on an Aperio AT2 slide scanner.
Colour-mapping fluorescence to virtual haematoxylin and eosin (vH&E)
A two-staged approach was used to convert original MUSE fluorescence images to an H&E-stained brightfield appearance. The first step consisted of unmixing the RGB colour image based on the (3-channel) spectral properties of the fluorescent dyes, using end-members selected from regions corresponding to cytoplasm and nuclei, with spectral correction to estimate the colour coordinates of the pure dye components. The second step converted the corresponding dye-specific abundance images into simulated H&E concentrations using a Beer-Lambert physical model of transillumination microscopy, wherein the absorption RGB values of haematoxylin and eosin are used to determine the colours in the computed output image. Additional details are provided in Fig. S4 and Supplementary Note 3.
Immunohistochemical and fluorescence in situ hybridization methods
Standard histological and immunohistochemical (IHC) procedures, including semiquantitative analyses were performed as previously published 33. Representative sections were stained by immunohistochemical labeling using DAKO OMNIS automated immunohistochemistry system (Dako, Agilent Technologies, Carpinteria, CA) using the following antibodies: (1) an anti-ATRX (α thalassemia/mental retardation syndrome X-linked)) rabbit polyclonal antibody (cat. No. HPA001906, Sigma-Aldrich, a part of Millipore-Sigma, St. Louis, MO); (2) an anti–Ki-67 mouse monoclonal antibody (GA626, prediluted, Dako, Agilent Technologies, Carpinteria, CA). Fluorescence in-situ hybridization (FISH) testing for 1p19q co-deletion on interphase cells was performed on formalin-fixed paraffin embedded tissue (FFPE): A) FFPE only; B) FFPE after MUSE. The tumor cells that were labeled with 19q13 Spectrum Orange/19p13 Spectrum Green using Vysis LSI 1p36/1q25 and LSI 19q13/19p13 dual-colour probes using a previously published protocol 34. Results are shown in Fig. S7.
RNA-Seq
To investigate whether MUSE staining and imaging, steps that involve the use of intercalating dyes and UV excitation, might affect labile RNA sample integrity, two core needle “biopsies”—obtained ex-vivo from a resected specimen—were processed in parallel. One was stained with Hoechst and rhodamine for 10 seconds as described above, and imaged over several fields for a total imaging time of about 2 minutes. The unstained and unimaged paired specimen was kept moist in PBS at room temperature during this time. After MUSE imaging, both cores were snap-frozen in liquid N2 and processed for RNA sequencing. Total RNA was isolated using RNeasy Mini Kit (Qiagen, Inc.). Subsequently, RNA quantity and quality were assessed on a NanoDrop spectrophotometer and an Agilent 2100 Bioanalyzer. Whole transcriptome profiling was performed using a directional, strand-specific mRNA-Seq approach in the UC Davis Genomics Shared Resource (GSR), and indexed RNA-Seq libraries were prepared. Double-stranded cDNA was generated as described 35,36, and libraries were sequenced on an Illumina MiSeq System (75-bp, paired-end; ~30 million reads/sample) as described 37.
To compare the results from the MUSE-imaged and control RNA sequencing runs, de-multiplexed raw sequence data (FASTQ) were aligned to the human genome (GRCh37) using the DRAGEN™ bioinformatics processor. The aligned reads (bam format) were analyzed using the RSeQC RNA-Seq Quality Control package 38,39. None of the metrics examined showed appreciable differences between the MUSE and non-MUSE samples outside of expected RNA-seq library variation. Supplementary Table 1 and Supplementary Fig. 8c summarize a few key metrics. The results provide reassurance that the approach of using MUSE-imaged small specimens will not seriously impair downstream molecular diagnostic applications.
Assessment of diagnostic suitability of MUSE images
A. General surgical pathology specimens, MUSE vs. H&E
42 cases from the general surgical pathology service at UC Davis were selected for a comparison between diagnoses obtained via MUSE and H&E imaging. Excess material beyond that required for patient care was obtained under an UC Davis IRB exemption allowing study of anonymized tissue samples. The distribution was weighted towards ovary, kidney, colon, breast, lung, and prostate, along with a few other sites of origin. Out of the 42 cases, 10 were benign, and the balance was either frankly malignant or borderline. No effort was made to select text-book examples of common pathologies, and some of the cases posed considerable degree of diagnostic difficulty. 8 specimens were examined in the fresh state, and the remainder had been formalin-fixed for variable periods of time ranging from a few days to a few weeks. Each case was imaged via MUSE using 10-second staining with rhodamine and Hoechst, and multiple fields of view were captured at 10X magnification. Companion tissue was submitted for fixation (if required) and standard histological processing followed by H&E staining and whole-slide scanning at 20X. Representative fields reflecting both normal (if present), adjacent and diagnostic regions were selected and matched by one of the authors, and then presented to two experienced, board-certified pathologists, with only tissue of origin information being provided (i.e., no age, gender, medical history or procedure). Each reviewer saw approximate 190 images over the 42 cases (mean 4.5 images per case, range 2–11). One reviewer made diagnosis viewing only the MUSE images (in both fluorescence and vH&E mode), and similarly, the other viewed only the H&E images; no communication between the reviewers occurred. Their diagnoses, along with comments, if any, more or less verbatim, are presented in Supplemental Information.
B. Neuropathology specimens
A panel of board-certified/eligible practicing anatomic pathologists and neuropathologists conducted blinded analysis to assess diagnostic accuracy achievable with MUSE images (primary MUSE and virtual H&E) when compared with diagnosis obtained by review of the standard FFPE, microtome-sectioned and H&E-stained glass slides of the same brain tumor specimens. Given the histopathological heterogeneity of CNS tumors and the fact that some specimens may contain a mixture of normal and lesional areas of interest, 2–4 representative images were captured from each case and selected by an independent board-certified neuropathologist in order to best represent diagnostic features. 24 adult patients (14 male, 10 female), mean age 54 years (range 19 – 83 years), who underwent surgical resection for newly diagnosed brain and spinal cord tumors were included in the study. 7 (29%) were diagnosed with diffuse astrocytic or oligodendroglial tumors; 8 (33.5%) with meningiomas; 3 (12.5%) with ependymal and choroid plexus tumors; 3 (12.5%) with tumors of the cranial or paraspinal nerves and 3 (12.5%) with metastatic tumors by the conventional methodology.
Each pair of test images (primary MUSE and vH&E) was accompanied by a questionnaire that included a series of potential diagnoses that included the above diagnoses. The pathologists had to pick the one answer that represented the correct diagnostic category.
Any differences in diagnosis between primary MUSE or vH&E as compared to the ground-truth diagnoses made by conventional methodology were noted as a major, clinically significant difference (false diagnosis/subclassification of tumor), as presented in Table S2.
Code availability
The Python code and executables to convert full-colour MUSE images to virtual H&E images are freely available on GitHub at https://github.com/UCDavisMUSE/colourmapper.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information.
Supplementary Material
Acknowledgments
We would like to acknowledge Ananya Datta-Mitra, M.D., Trevor McBroom, Adebimpe Adelaja, Alex Krueger, and Lauren Martinez for helping with sample preparation and imaging; John Wilson and Douglas Peabody for assisting with tissue procurement; and Elizabeth Hillman, Ph.D. for providing critical feedback. This work was partially supported by UC Davis Department of Pathology and Laboratory Medicine start-up funds, a UC Davis Science Translation and Innovative Research (STAIR) Grant and an unrestricted gift from Agilent Technologies. This work was performed in part under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344.
Footnotes
Author contributions
F.F., R.L. and S.D. developed the original observations of tissue autofluorescence in the ultraviolet; F.F. and R.L. designed and fabricated the MUSE microscope based on an original design by S.D. and conducted wavelength-dependent depth measurements. R.L., F.F., M.T., A.T., J.K. performed experiments on sample preparation, staining, and comparison to traditional histology. R.L., M.T., J.K., and M.L. designed and performed the comparison of MUSE and traditional histology in CNS cases. R.L., A.T., designed and performed the comparison of MUSE tissues to traditional histology, which were viewed and interpreted by J.B. and A.B. R.L., F.F., M.L. and J.M. designed and performed the FISH and RNA-seq experiments. All authors discussed and interpreted the results, wrote and edited the paper.
Competing interests
R.L. and S.D. are co-founders of a start-up company, Muse Microscopy Inc., which is involved in commercializing MUSE technology. The other authors declare no competing financial interests.
References
- 1.Rastogi V, et al. Artefacts: a diagnostic dilemma - a review. J Clin Diagn Res. 2013;7:2408–2413. doi: 10.7860/JCDR/2013/6170.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neil MA, Juskaitis R, Wilson T. Method of obtaining optical sectioning by using structured light in a conventional microscope. Opt Lett. 1997;22:1905–1907. doi: 10.1364/ol.22.001905. [DOI] [PubMed] [Google Scholar]
- 3.Dobbs J, et al. Confocal fluorescence microscopy for rapid evaluation of invasive tumor cellularity of inflammatory breast carcinoma core needle biopsies. Breast Cancer Res Treat. 2015;149:303–310. doi: 10.1007/s10549-014-3182-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tao YK, et al. Assessment of breast pathologies using nonlinear microscopy. Proc Natl Acad Sci U S A. 2014;111:15304–15309. doi: 10.1073/pnas.1416955111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang D, et al. Endoscopic probe optics for spectrally encoded confocal microscopy. Biomedical optics express. 2013;4:1925–1936. doi: 10.1364/BOE.4.001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orringer DA, et al. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nature Biomedical Engineering. 1:0027. doi: 10.1038/s41551-016-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu JT, et al. Efficient rejection of scattered light enables deep optical sectioning in turbid media with low-numerical-aperture optics in a dual-axis confocal architecture. Journal of biomedical optics. 2008;13 doi: 10.1117/1.2939428. 034020-034020-034011. [DOI] [PubMed] [Google Scholar]
- 8.Bouchard MB, et al. Swept confocally-aligned planar excitation (SCAPE) microscopy for high-speed volumetric imaging of behaving organisms. Nat Photonics. 2015;9:113–119. doi: 10.1038/nphoton.2014.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gabriele ML, et al. Optical coherence tomography: history, current status, and laboratory work. Invest Ophthalmol Vis Sci. 2011;52:2425–2436. doi: 10.1167/iovs.10-6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin B, Urayama S, Saroufeem RM, Matthews DL, Demos SG. Real-time microscopic imaging of esophageal epithelial disease with autofluorescence under ultraviolet excitation. Optics express. 2009;17:12502–12509. doi: 10.1364/oe.17.012502. [DOI] [PubMed] [Google Scholar]
- 11.Lin B, Urayama S, Saroufeem RM, Matthews DL, Demos SG. Characterizing the origin of autofluorescence in human esophageal epithelium under ultraviolet excitation. Optics express. 2010;18:21074–21082. doi: 10.1364/OE.18.021074. [DOI] [PubMed] [Google Scholar]
- 12.Kather JN, et al. New Colours for Histology: Optimized Bivariate Colour Maps Increase Perceptual Contrast in Histological Images. PloS one. 2015;10:e0145572. doi: 10.1371/journal.pone.0145572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fereidouni F, Datta-Mitra A, Demos S, Levenson R. SPIE BiOS. International Society for Optics and Photonics; 93180F-93180F-93186. [Google Scholar]
- 14.Ho D, Fereidouni F, Levenson RM, Jagdeo J. Real-time, High-resolution, In Vivo Characterization of Superficial Skin With Microscopy Using Ultraviolet Surface Excitation (MUSE) JOURNAL OF DRUGS IN DERMATOLOGY. 2016;15:1344–1346. [PMC free article] [PubMed] [Google Scholar]
- 15.Zeskind BJ, et al. Nucleic acid and protein mass mapping by live-cell deep-ultraviolet microscopy. Nature Methods. 2007;4:567–569. doi: 10.1038/nmeth1053. [DOI] [PubMed] [Google Scholar]
- 16.Zaak D, et al. Ultraviolet-excited (308 nm) autofluorescence for bladder cancer detection. Urology. 2002;60:1029–1033. doi: 10.1016/s0090-4295(02)01999-4. [DOI] [PubMed] [Google Scholar]
- 17.Ashutosh S, Stephen G. John Wiley & Sons, Inc; New York: 1999. [Google Scholar]
- 18.Kenny KB. 8,639,013 B2. US Patent US. 2011
- 19.Garsha K, et al. Google Patents. 2016. [Google Scholar]
- 20.Hui P, Buza N. Atlas of intraoperative frozen section diagnosis in gynecologic pathology. Springer; 2015. [Google Scholar]
- 21.Foschini MP, Baldovini C, Ishikawa Y, Eusebi V. The value of large sections in surgical pathology. Int J Breast Cancer. 2012;2012 doi: 10.1155/2012/785947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abels E, Pantanowitz L. Current state of the regulatory trajectory for whole slide imaging devices in the USA. J Pathol Inform. 2017;8:23–23. doi: 10.4103/jpi.jpi_11_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adeyi OA. Pathology services in developing countries-the West African experience. Arch Pathol Lab Med. 2011;135:183–186. doi: 10.1043/2008-0432-CCR.1. [DOI] [PubMed] [Google Scholar]
- 24.Benediktsson H, Whitelaw J, Roy I. Pathology services in developing countries: a challenge. Arch Pathol Lab Med. 2007;131:1636–1639. doi: 10.1043/1543-2165(2007)131[1636:PSIDCA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 25.Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961–1971. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapp JR, et al. Variation in pre-PCR processing of FFPE samples leads to discrepancies in BRAF and EGFR mutation detection: a diagnostic RING trial. J Clin Pathol. 2015;68:111–118. doi: 10.1136/jclinpath-2014-202644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penland SK, et al. RNA expression analysis of formalin-fixed paraffin-embedded tumors. Lab Invest. 2007;87:383–391. doi: 10.1038/labinvest.3700529. [DOI] [PubMed] [Google Scholar]
- 28.Maes E, et al. Analysis of the formalin-fixed paraffin-embedded tissue proteome: pitfalls, challenges, and future prospectives. Amino Acids. 2013;45:205–218. doi: 10.1007/s00726-013-1494-0. [DOI] [PubMed] [Google Scholar]
- 29.Gnanapragasam VJ. Unlocking the molecular archive: the emerging use of formalin-fixed paraffin-embedded tissue for biomarker research in urological cancer. BJU international. 2010;105:274–278. doi: 10.1111/j.1464-410X.2009.08665.x. [DOI] [PubMed] [Google Scholar]
- 30.Ragan T, et al. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat Methods. 2012;9:255–258. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hapke MR, Dehner LP. The optically clear nucleus. A reliable sign of papillary carcinoma of the thyroid? Am J Surg Pathol. 1979;3:31–38. doi: 10.1097/00000478-197902000-00004. [DOI] [PubMed] [Google Scholar]
- 32.Lakowicz JR. Principles of fluorescence spectroscopy. Springer Science & Business Media; 2013. [Google Scholar]
- 33.Lechpammer M, et al. Pathology of inherited manganese transporter deficiency. Annals of neurology. 2014;75:608–612. doi: 10.1002/ana.24131. [DOI] [PubMed] [Google Scholar]
- 34.Woehrer A, et al. FISH-based detection of 1p 19q codeletion in oligodendroglial tumors: procedures and protocols for neuropathological practice-a publication under the auspices of the Research Committee of the European Confederation of Neuropathological Societies (Euro-CNS) Clinical neuropathology. 2010;30:47–55. doi: 10.5414/npp30047. [DOI] [PubMed] [Google Scholar]
- 35.Borodina T, Adjaye J, Sultan M. A strand-specific library preparation protocol for RNA sequencing. Methods Enzymol. 2011;500:79–98. doi: 10.1016/B978-0-12-385118-5.00005-0. [DOI] [PubMed] [Google Scholar]
- 36.Levin JZ, et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 39.Wang L, et al. Measure transcript integrity using RNA-seq data. BMC Bioinformatics. 2016;17:58. doi: 10.1186/s12859-016-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information.