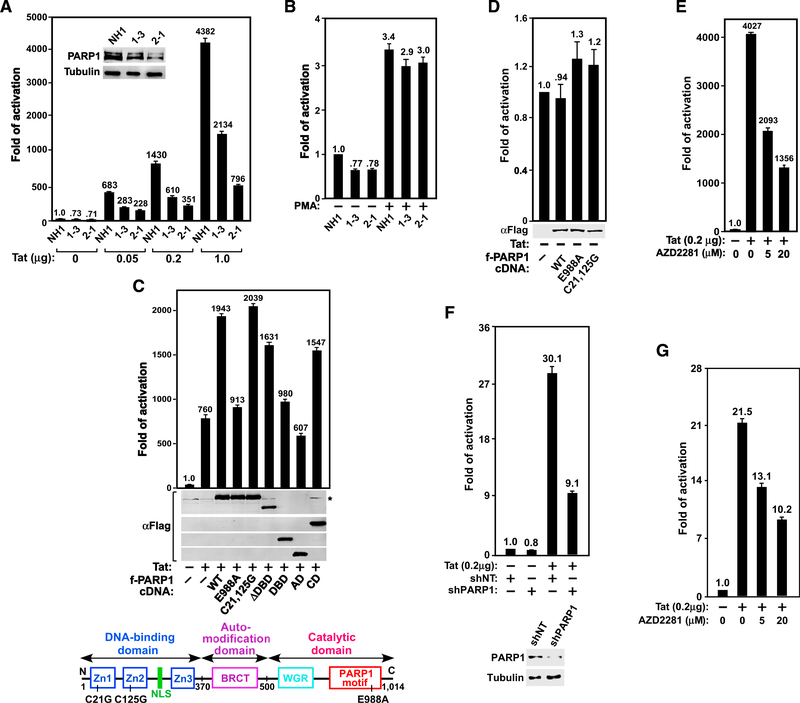
Figure 1. PARP1 and Its Catalytic Activity Are Required for Tat- but Not PMA-Induced Activation of HIV-1 Transcription.
(A and B) The HeLa-based NH1 cells with an integrated HIV-1 LTR-luciferase reporter construct and two NH1-derived PARP1 KD clones 1–3 and 2–1 were transfected with the indicated amounts of Tat cDNA (A) or treated with 200 nM PMA for 24 hr (B). Luciferase activities were measured in these cells and compared with that in Tat-free (A) or untreated NH1 cells (B), which was set to 1. The inset in (A) shows PARP1 levels in the three cell lines as revealed by western blotting (WB). (C and D) The indicated Flag-tagged PARP1 mutants were co-expressed with (C) or without (D) Tat in 2–1 cells and examined using anti-Flag WB. Luciferase activities were analyzed as in (A). The domain structure of PARP1 and point mutations are shown in (C).
(E and G) NH1 (E) or the Jurkat-based 1G5 cells containing an integrated HIV-1 LTR-luciferase reporter construct (G) were transfected with or without the Tat cDNA and then treated with the indicated concentrations of AZD2281. Luciferase activities were measured and analyzed as in (A).
(F) 1G5 cells were stably transfected with plasmids expressing the indicated shRNA (NT, non-target) and/or the Tat cDNA. Also shown are PARP1 levels in the KD cells. Luciferase activities were measured and analyzed as in (A).
Error bars in all graphs represent mean ± SD from three separate measurements. See also Figure S1