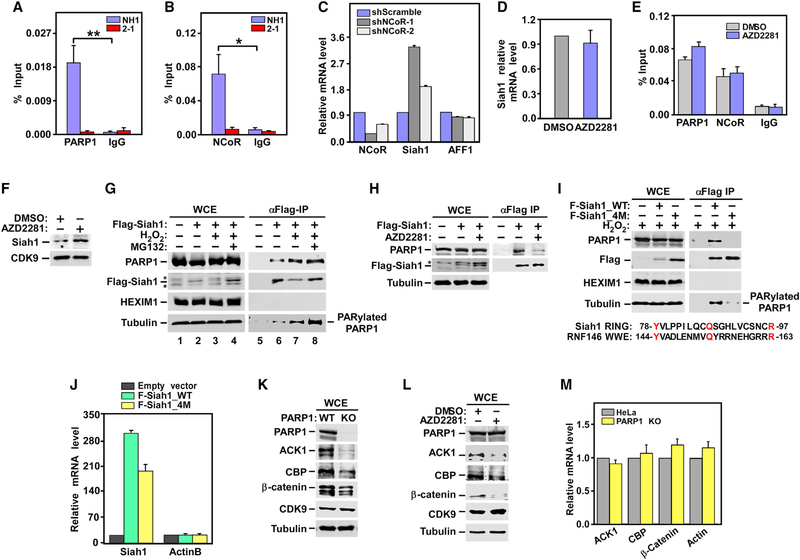
Figure 4. Mechanisms by which PARP1 Suppresses Siah1 Expression at Transcriptional and Protein Stability Levels: The Effect of PARP1 Depletion or Inhibition on Protein and mRNA Levels of Three Other Siah1 Substrates.
(A, B, and E) NH1 and the NH1-based PARP1 KD line 2–1 (A and B) or HeLa cells treated with DMSO or AZD2281 (E) were analyzed by ChIP-qPCR for the bindings of PARP1 and NCoR to the Siah1 gene promoter, with total mouse or rabbit IgG used as a negative control. The ChIP-qPCR signals were normalized to those of input DNA. Statistical significance was calculated from two-tailed Student’s t-test. *p < 0.05, **p < 0.005.
(C) Total RNAs isolated from HeLa cells expressing the indicated shRNAs were analyzed using qRT-PCR for the genes marked at the bottom. The signals were normalized to those of GAPDH and displayed.
(D and F) HeLa cells were treated with DMSO or AZD2281. Total RNAs were analyzed using qRT-PCR for the relative levels of Siah1 transcripts, which were normalized to those of GAPDH and shown (D). NEs were examined using WB for the indicated proteins (F).
(G and H) HeLa cells expressing WT Flag-Siah1 were treated with H2O2, pre-treated with MG132 and then followed by H2O2 (G), or treated with AZD2281 (H). WCE and anti-Flag immunoprecipitates (IPs) derived from WCE were examined using WB. Asterisks denote the positions of non-specific bands.
(I) HeLa cells were transfected with the same amount of plasmid expressing WT or the 4M mutant Flag-Siah1. WCE and anti-Flag IP derived from WCE wereexamined using WB. Also shown is an alignment between the RING domain of Siah1 and WWE domain of RNF146.
(J) Total RNAs isolated from HeLa cells either untransfected or transfected with the indicated expression constructs were analyzed using qRT-PCR as in (C).
(K and L) WCE from WT, the PARP1 KO (K), or the AZD2281-treated HeLa cells (L) were examined using WB for the indicated proteins.
(M) Total RNAs isolated from either WT or the PARP1 KO HeLa cells were analyzed using qRT-PCR as in (C)
The error bars in panels (A)–(E), (J), and (M) represent mean ± SD from three independent measurements. See also Figures S2–S4.