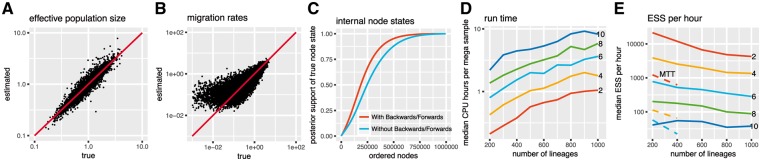
Fig. 2.
Inference of effective population sizes, migration rates and node states. (A) Inferred effective population sizes on the y-axis versus true effective population sizes on the x-axis. The effective population sizes for the tree simulations were sampled from a lognormal (-0.125, 0.5) distribution. The coverage of migration rate estimates was 95.5% and for effective population size estimates 94.9%. (B) Inferred migration rates on the y-axis versus true migration rates on the x-axis. The migration rates between states were sampled from an exponential distribution with mean = 0.5. (C) Inferred node states using MASCOT with and without the backwards/forwards algorithm. (D) Median CPU time per mega sample depending on the number of lineages and the number of different states. The CPU time was taken from 100 replicates of the simulation scenario used. (E) Median posterior ESS per hour from 100 replicates from MASCOT and MultiTypeTree (dashed lines). The different colours indicate the different number of states. Dashed lines show median ESS per hour values for MultiTypeTree