Fig. 1.
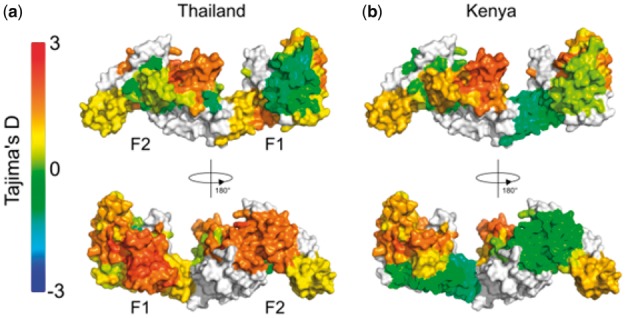
Tajima’s D calculation applied as a 3D sliding window over the protein structure of P. falciparum EBA-175 RII. The F1 and F2 domains are indicated on the monomeric structure. Nucleotide sequences were obtained from P. falciparum isolates from (a) Thailand (n = 48) and (b) Kenya (n = 39) (Verra et al., 2006). The BioStructMap Python package was used to apply Tajima’s D calculations using a 3D sliding window with a radius of 15 Å. The structural model is available via ModBase, accession number: ed998157a605f5e58ed66e198e0ae1ab. Structures were visualized with PyMOL
