Fig. 3.
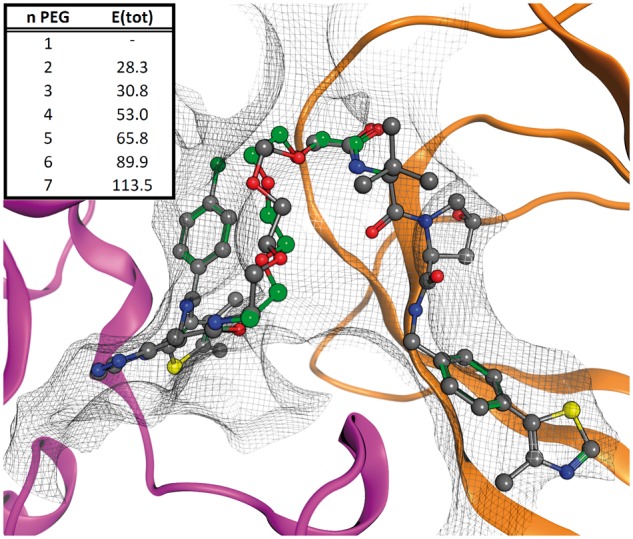
Computational assessment of spacer length for BRD4-VHL bivalent ligand. The tool was used to test a database 1- to 7-unit (n) PEG spacers. The energy of the system (E(tot), kcal/mol) shows the best results use two and three PEG repeats. Both correspond to experimentally valid linkers. The computationally derived three-unit PEG structure from the tool (green) superposes excellently to the X-ray structure (grey)
