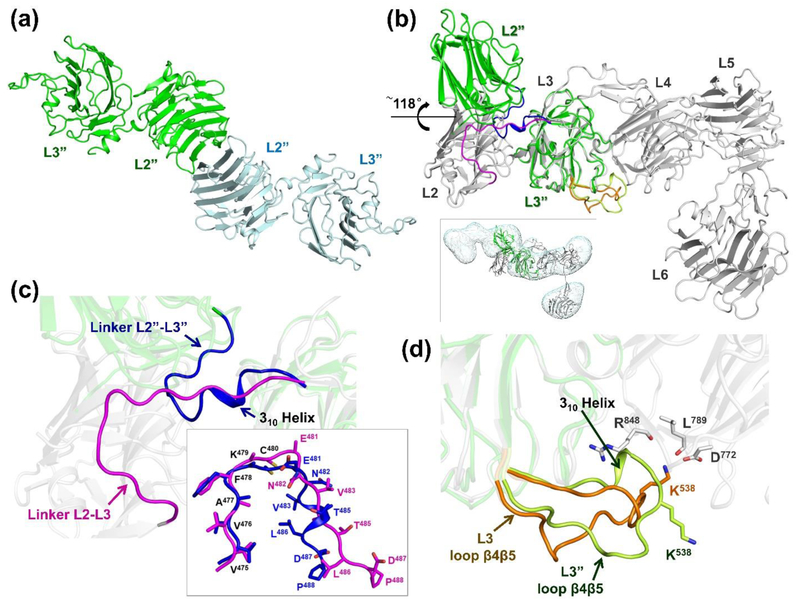
Figure 6: Crystal structure of n1α L2-L3.
a) Cartoon diagram of the two n1α L2-L3 tandems in the asymmetric unit.; b) Superposition of n1α L2-L3 (green) on n1α (grey; pdbid:3QCW). Inset shows n1α L2-L3 docked in the IPET map of a n1α L1-L6 particle with a bent central core, shown as dual iso-surfaces contoured at volumes corresponding to 1.2 time the molecular mass of ~141 kDa.; c) Close-up of the interface between L2 and L3 shown in b). The linker between L2 and L3 undergoes a dramatic movement in n1α L2-L3 (blue; linker L2”-L3”) compared to its counterpart in the n1α ectodomain (magenta; linker L2-L3 from pdbid:3QCW). Inset shows the superposition of these linkers; d) Close up of the interface between L3 and L4 shown in b). Loopβ4-β5 in L3 as seen in the n1α L2-L3 fragment (light green) and its counterpart as seen in n1α (orange; pdbid:3QCW). The Ca2+-binding site of L4 is formed by the side chain of D772 and the backbone carbonyls of R848 and L789.