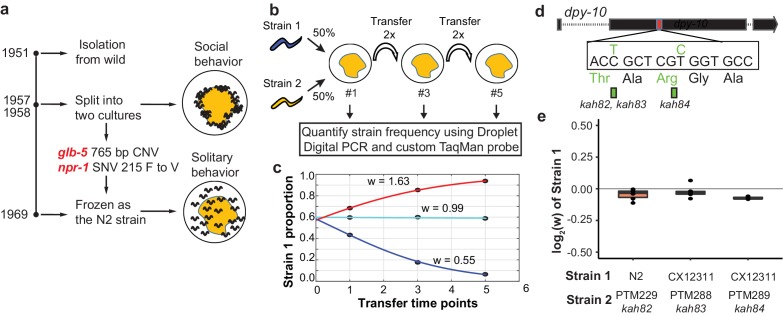
Figure 1. Schematic of competition assays used to measure relative fitness levels between two strains.
(a) Overview of life history of the standard reference N2 strain since its isolation from the wild. Derived alleles in npr-1 and glb-5 arose and fixed after 1957 and before 1969 when methods for cryopreservation were developed. These two alleles were identified for their role in changing foraging behavior on bacterial lawns from social to solitary behavior. (b) Schematic of pairwise competition experiments used throughout the paper to quantify fitness differences between two strains. (c) Relative proportion of each strain as ascertained by Droplet Digital PCR using a custom TaqMan probe (dots) is used to estimate the relative fitness between the two strains (line). (d) Silent mutations were edited into the 90th or 92nd amino acid of the dpy-10 gene using CRISPR/Cas9 to create a common SNV for Droplet Digital PCR. We refer to these as barcoded strains. (e) Competition experiments between the parent strain (top) and the same strain containing one of the silent mutations. We display the result from each competition experiment as a single dot overlaid on top of a boxplot showing the mean, first, and third quartiles of all replicates.